E3 ligase ligands are molecules that can specifically bind to E3 ubiquitin ligases.
Mechanism of Action of E3 Ligase Ligands
Inducing Target Protein Ubiquitination and Degradation
E3 ligase ligands selectively attach to E3 ubiquitin ligases which allows these ligases to identify and bind to their target proteins. Ubiquitin becomes activated initially by E1 ubiquitin-activating enzyme before it joins E2 ubiquitin-conjugating enzyme and is ultimately conjugated to target protein lysine residue through E3 ligase catalysis. The effectiveness and target precision of this reaction improves when the E3 ligase ligand is present and polyubiquitin chains form through successive attachment of ubiquitin molecules to the protein. The proteasome recognizes and degrades target proteins once they bind with a K48 polyubiquitin chain which serves to maintain cellular protein balance.
Regulating the Activity of E3 Ligases
Conformational Change: The binding of the ligand to the E3 ligase triggers conformational changes which either enhance access of the active site to the target protein or boost the E3 ligase's functionality. Certain small molecule ligands augment the catalytic function of E3 ligases by maintaining their active conformation.
Promoting E3 Ligase Auto-Ubiquitination: E3 ligases that possess auto-ubiquitination capabilities experience enhanced auto-ubiquitination when ligands bind to them. The activity levels and stability of E3 ligases remain under control through their auto-ubiquitination process which also affects their protein interaction capabilities. RING1B is an example of an E3 ligase that self-induces ubiquitin chain formation through auto-ubiquitination which activates its ability to ubiquitinate substrates.
Participating in Protein Complex Formation
E3 ligase ligands serve as connectors between E3 ligases and other proteins or protein complexes which results in the formation of larger synergistically functioning protein complexes. E3 ligase ligands recruit essential signaling proteins to the E3 ligase to create a signaling complex that regulates protein ubiquitination and degradation more effectively in the signaling pathway thus altering cellular physiological functions.
Regulating Intracellular Signaling Pathways
Regulating Transcription Factor Activity: Ubiquitination modifications play a regulatory role in the function of many transcription factors. The levels of ubiquitination of transcription factors controlled by E3 ligase ligands determine their nuclear accumulation and transcriptional activity which then influences gene expression patterns. Some ligands lead to the breakdown of transcription factors which results in inhibited gene expression while different ligands can stabilize transcription factors which enhances gene transcription.
Affecting Key Node Proteins in Signaling Pathways: Multiple cellular signaling pathways depend on key node proteins which control signal transmission through their functional activity and stable interactions. E3 ligase ligands function on node proteins and control their ubiquitination modifications which affects the signaling pathway's activity. The TNF-α signaling pathway demonstrates how E3 ligase ligands control the ubiquitination of IKK complex proteins which results in changes to NF-κB signaling pathway activation.
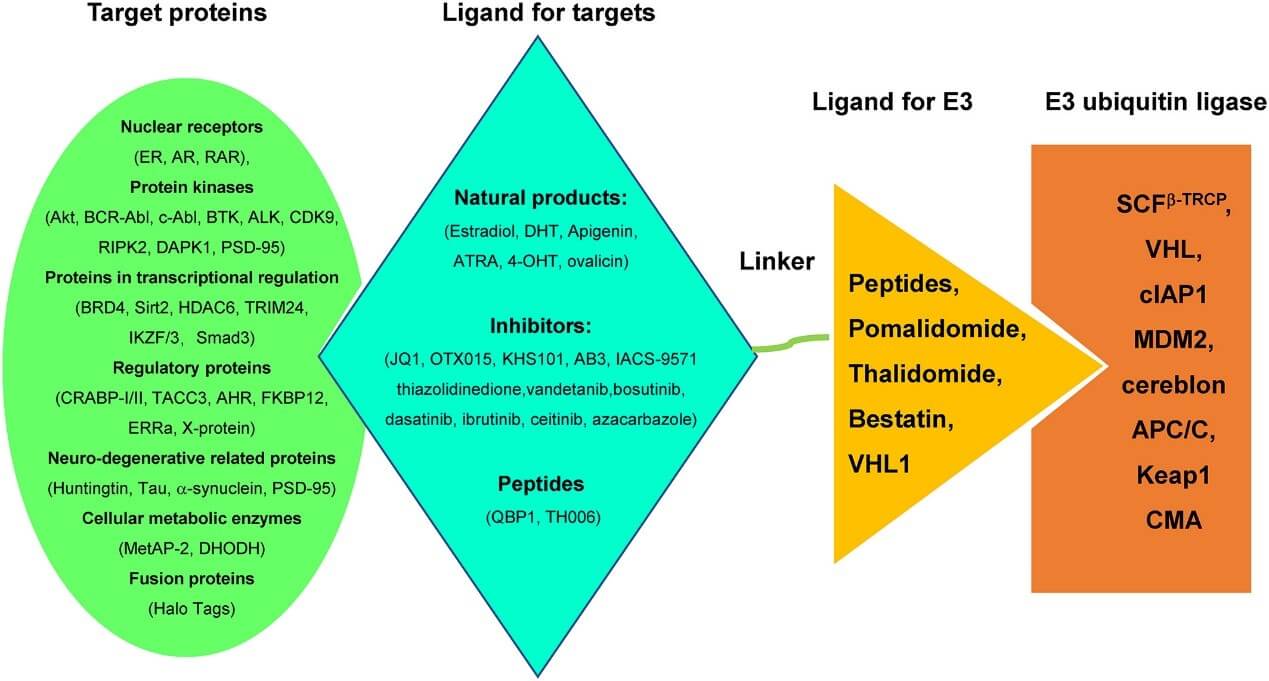
Common Types of E3 Ligase Ligands
CRBN Ligands
Description: CRBN is the substrate recognition subunit of the CRL4CRBN E3 ligase complex. IMiD drugs like thalidomide , lenalidomide , and pomalidomide are common CRBN ligands. Thalidomide was originally developed as a sedative for pregnant women to treat morning sickness but was withdrawn from the market due to teratogenic effects. However, it was later repurposed as an immunomodulatory drug, and its target protein was identified as CRBN. Lenalidomide and pomalidomide also target CRBN and play significant roles in the treatment of diseases like multiple myeloma.
Application: By binding to CRBN, these ligands alter CRBN's ability to recognize substrate proteins, enabling the degradation of transcription factors like Ikaros and Aiolos, offering potential for treating hematologic malignancies and other diseases.
VHL Ligands
Description: VHL is the substrate recognition subunit of the Cullin 2 RING-VHL E3 ligase complex. VHL ligands, such as VH032-cyclopropane-F, are used in PROTAC design. VHL plays an essential role in cellular processes such as oxygen sensing and tumor suppression. VHL ligands help recruit the VHL protein to target proteins for recognition and ubiquitination by the E3 ligase.
Application: These ligands are useful for degrading tumor-associated proteins like HIF-1α, with significant potential in cancer treatment.
IAP Ligands
Description: The Inhibitor of Apoptosis Proteins (IAP) family includes c-IAP1, c-IAP2, and XIAP. IAP ligands like MV1, bestatin-methyl ester (ME-BS), and LCL-161 bind to the BIR domains of IAP proteins, inducing self-ubiquitination and degradation of IAP proteins.
Application: Degrading IAP proteins can activate TNF signaling pathway-dependent cell death, showing potential for cancer treatment.
MDM2 Ligands
Description: MDM2 is an E3 ubiquitin ligase that regulates the ubiquitination and degradation of p53. Nutlin-3 and RG7388 are common MDM2 ligands. Nutlin-3 binds to MDM2, preventing its interaction with p53, thereby stabilizing p53 protein.
Application: These ligands can be used to treat p53 wild-type tumors by inhibiting MDM2 activity and activating the tumor-suppressive functions of p53.
DCAF Ligands
Description: The DCAF protein family is a substrate recognition subunit of the Cull4 E3 ligase complex. Sulfonamide derivatives like indisulam, E7820, and chloroquine-quinoline sulfonamide act as DCAF ligands, inducing protein-protein interactions between the E3 ligase and target proteins.
Application: These ligands can be used to design PROTACs targeting proteins like BRD9 and BTK for degradation, with potential applications in cancer treatment and other diseases.
RNF Ligands
Description: RNF family proteins, such as RNF4 and RNF114, are E3 ubiquitin ligases. CCW16 has high affinity for RNF4, and Nimbolide is a natural product identified as a covalent ligand for RNF114.
Application: These ligands can be used in research and treatment of diseases related to RNF family proteins.
AhR Ligands
Description: The Aryl Hydrocarbon Receptor (AhR) is a ligand-activated transcription factor and a component of E3 ligase complexes. β-NF is an AhR ligand that binds to AhR, activating the AhR signaling pathway.
Application: These ligands can be used to design PROTACs targeting proteins like CRABPs for degradation, showing potential for treating related diseases.
FEM1B Ligands
Description: FEM1B is the substrate recognition subunit of the CUL2 E3 ligase complex. EN106, a chloroacetamide-based covalent ligand, binds to FEM1B.
Application: These ligands can be used in research and treatment of diseases related to FEM1B.
KEAP1 Ligands
Description: KEAP1 is the substrate recognition subunit of the Cull3 E3 ligase complex and interacts with Nrf2 to regulate cellular protective proteins. RTA402 and methyl bardoxolone (CDO-Me) are KEAP1 ligands that bind to KEAP1 and influence Nrf2 activity.
Application: These ligands can be used to treat diseases associated with oxidative stress and inflammation.
Research Progress and Applications
Application in PROTAC Technology: E3 ligase ligands are a critical component of PROTAC (Proteolysis Targeting Chimeras) molecules, which consist of a target protein ligand, a linker, and an E3 ligase ligand. By carefully designing and selecting E3 ligase ligands, efficient degradation of target proteins can be achieved, providing a new avenue for drug development.
New Drug Development: With the deepening research on E3 ligase ligands, some PROTAC molecules based on E3 ligase ligands have entered clinical trials. For example, VHL-based PROTACs are being explored for the treatment of cancers and other diseases.
How Do E3 Ligase Ligands Influence Target Protein Degradation?
In cells, protein degradation primarily occurs through the ubiquitin-proteasome system (UPS). E3 ligase ligands influence target protein degradation by participating in this system. The specific mechanisms are as follows:
Binding and Recruitment
E3 ligase ligands bind specifically to E3 ubiquitin ligases, effectively "equipping" the E3 ligase with a "localization device," allowing it to accurately recognize and approach the target protein. For instance, in PROTAC technology, one end of the molecule is a target protein ligand, and the other end is an E3 ligase ligand. These two are connected by a linker. Once the PROTAC molecule enters the cell, the E3 ligase ligand binds to the E3 ligase, while the target protein ligand binds to the target protein, bringing the E3 ligase and the target protein together to form a "target protein - PROTAC - E3 ubiquitin ligase" ternary complex.
Ubiquitination Marking
Under the action of E3 ligases, ubiquitin molecules are transferred to the lysine residues on the target protein, forming a polyubiquitin chain. This process acts like a "degradation tag" for the target protein, marking it for recognition by the proteasome within the cell. Typically, when the target protein is conjugated with a K48-linked polyubiquitin chain, it is specifically recognized by the 26S proteasome.
Proteasomal Degradation
The ubiquitinated target protein is subsequently captured and degraded by the 26S proteasome. The 26S proteasome consists of the 19S regulatory particle and the 20S core particle. The 19S regulatory particle is responsible for recognizing the ubiquitinated target protein and deubiquitinating it. The target protein then enters the 20S core particle, where it is cleaved into short peptide fragments. These peptide fragments are ultimately degraded into amino acids, which are reused in cellular metabolic processes.
Regulation of Signaling Pathways
By influencing the degradation of target proteins, E3 ligase ligands can regulate intracellular signaling pathways. For example, in tumor cells, certain E3 ligase ligands promote the degradation of oncogenic proteins, inhibiting signaling pathways that drive tumor cell proliferation and survival, thereby exerting antitumor effects. In neurodegenerative diseases, E3 ligase ligands can target and degrade toxic proteins associated with the disease, helping to balance cellular signaling and slow disease progression.











![3-(1-oxo-3,5,6,7-tetrahydropyrrolo[3,4-f]isoindol-2-yl)piperidine-2,6-dione](https://resource.bocsci.com/structure/2616539-03-6.gif)










