PROTAC building blocks have left the confines of medicinal chemistry and are now benchtop staples in academic labs as diverse as plant biology and polymer science. The same heterobifunctional platform that targets an E3 to a disease target can be repurposed to traffic transcription factors to chloroplasts, to deplete antifungal resistance proteins in crop pathogens, or to create self-immolative biomaterials whose mechanical properties are gated by a user-defined protein trigger. Because pomalidomide-derived CRBN recruiters are commercially available in gram quantities with pre-installed click handles, non-medicinal groups can access catalytic protein knock-down without synthesizing a single milligram of new chemistry, turning a specialized drug-discovery tool into a broadly-accessible biological switch that is as routine as CRISPR or siRNA.
Introduction-The Expanding Utility of PROTAC Chemistry
The idea of induced proximity has leapt beyond therapeutic degradation and been adopted wherever a protein needs to be erased, removed or post-translationally rewired. PROTAC chemistry provides a modular platform – one ligand for the protein, one ligand for the cellular machinery and a variable spacer – so that any lab with the skills to manipulate small molecules can in principle hijack endogenous pathways without genetic modification. As a result, the same building blocks that medicinal chemists order for cancer programmes are being repurposed by materials scientists who want to dissolve a proteinaceous interphase, or by synthetic biologists who need to pulse-deplete a signalling node in a way that is both dose-dependent and reversible.
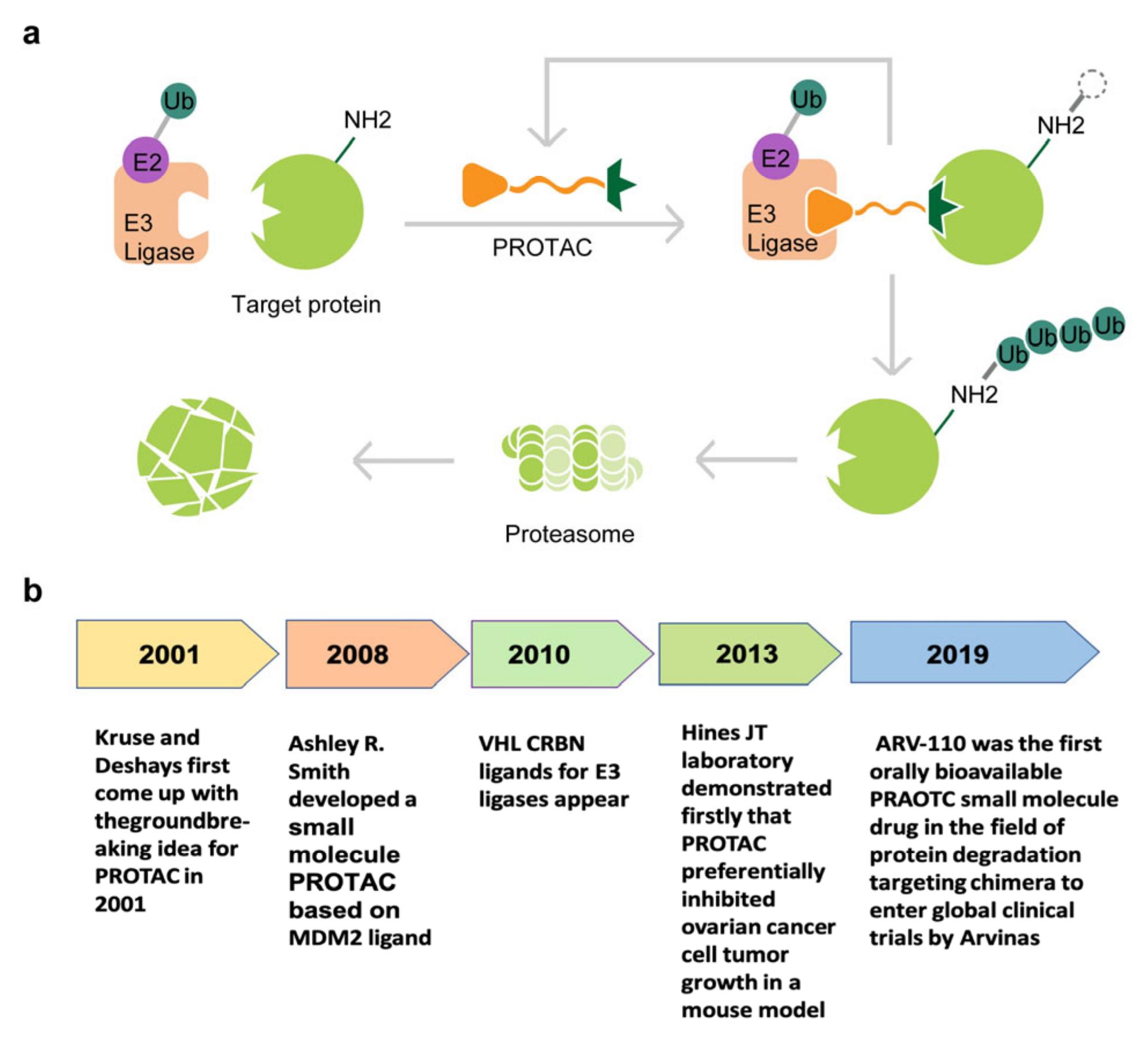 Fig. 1 The structure and development process of protac: (a) The structure of PROTAC and the mechanism by which it targets protein degradation; (b) Major events in the history of PROTAC.1,6
Fig. 1 The structure and development process of protac: (a) The structure of PROTAC and the mechanism by which it targets protein degradation; (b) Major events in the history of PROTAC.1,6
How PROTAC Principles Are Influencing Broader Research Fields
PROTAC logic—proximity-induced ubiquitination followed by irreversible removal—has been adopted into fields that have historically not been focused on drug discovery. Plant biologists have adopted the idea into “degron walks” that are used to map transcription-factor redundancy during abiotic stress; by fusing a small molecule ligand to auxin-responsive F-box proteins, one can target a chosen TF for degradation within minutes of watering seedlings with a cognate degrader, thereby bypassing lethality that occurs with constitutive knock-out lines. In materials science, ternary-complex formation is used as a supramolecular cross-link: a bifunctional small molecule transiently bridges a surface-tethered E3 ligase to a target protein that is adsorbed on a gold chip, which results in on-chip ubiquitination and subsequent proteasome-mediated release and resetting of the biosensor for multiple analyte captures. Neuroengineers have even combined optogenetics with photocaged PROTACs to enable light-directed degradation of synaptic scaffolds, allowing one to map the molecular circuits of memory consolidation with millisecond resolution, which is impossible with genetic approaches. In astrochemistry, mineral surfaces on early Earth are hypothesized to have served as primitive linkers, adsorbing both substrate peptides and metal-ion ligases to begin prebiotic protein turnover—turning PROTAC chemistry into a potential protagonist in molecular evolution. Common to all of these is the denominator of event-driven irreversibility: instead of quantitating how much a protein does, one can watch what happens when it goes away, a paradigm shift that redefines causation in terms of correlation and disappearance.
Academic Interest in Protein Degradation Mechanisms
Universities have started to incorporate PROTACs into their instruction of ubiquitin biology, which previously required genetic model systems. For example, a common graduate course might have students synthesize a library of alkyl-linked chloroalkane-cRHH tandems; varying the linker by two-atom increments, they map a degradation profile against a Halo-tagged reporter and directly observe ubiquitin chain elongation on a Western blot, all in a single afternoon. The result is a replacement for weeks of CRISPR-based gene editing that distills into chemical genetics both the primary role of topology (rather than affinity) in ubiquitin transfer. Mechanistic biochemistry labs take advantage of PROTAC tunability to explore preference codes for specific lysines: Interchanging VHL and RNF4 ligase modules while holding the target binder constant, for example, illustrates how E2-Ub charging rate dominates over local primary sequence and answers a long-standing question of whether ubiquitination signals are contextual or deterministic. Finally, chemical biology research groups offer as a service "degradomics": in-situ PROTAC treatment followed by diGly remnant profiling can, in a single LC-MS run, map every ubiquitin acceptor site in a cell, a task impossible with genetic knockouts due to compensatory phosphorylation that obscures otherwise latent lysines. In the most telling sign of a maturing field, funding agencies have formed a separate peer-review panel to consider PROTAC-enabled target validation as its own category, no longer subsumed by the more general push for PROTACs to be applied as a pharmaceutical strategy; PROTAC chemical knock-out has become a pillar of ubiquitin biology itself, not just a complementary approach to classical genetics. The result is that the ubiquitin proteasome system is taught as a programmable language, and the academic chemist's role is to provide the vocabulary—warheads, linkers and ligases—while biologists write the sentences that living cells read as disappearance.
Table 1 Cross-Disciplinary Uses of PROTAC Building Blocks.
| Field | Target Class | Degrader Format | Scientific Question Addressed |
|---|
| Synthetic biology | Fluorescent reporters | Light-caged PROTAC | Protein half-life tuning in single cells |
| Materials science | Cross-linking enzymes | Hydrogel-embedded recruiter | Stiffness modulation in bio-scaffolds |
| Agriculture | Anti-nutritional grain | Feed-additive degrader | Nutrient bioavailability without GMO |
| Chemical biology | Orphan RNA-binding | RNA-PROTAC chimera | Ligandability of non-enzymatic proteins |
Applications in Academic Research
Repurposing pomalidomide-based PROTAC building blocks as general purpose molecular scalpels, academic laboratories are now applying them to functional genomics problems without recourse to transfection or genetic manipulation. The ubiquitin-proteasome system is evolutionarily conserved from yeast to mammalian cells, so the same hetero-bifunctional chemistries can be applied in cell lines, primary neurons, nematodes or even plant protoplasts to test whether a newly identified protein is required for a phenotype. Catalytic activity of the degrader means that only transient exposure is needed, so you can add the compound, wait for the loss of function phenotype, and then wash out the molecule to test for recovery of the phenotype, a sort of experimental choreography that would be impossible with CRISPR knock-outs. Availability of photocaged or pH-cleavable linkers even allows for spatiotemporal control, so that degradation can be limited to a single organelle, or induced in response to a physiological cue such as neuronal depolarization or immune-cell activation.
Studying Ubiquitination and Proteasome Pathways
PROTACs have an in-built, chemically-encoded timer that starts when the heterobifunctional molecule is added and stops when it is washed out. This converts the kinetics of ubiquitination from being measured on the hour scale, as for heat-shock or proteasome inhibition, to the minute scale. By adjusting linker length between the pomalidomide head and the warhead, students can record live-cell movies of ternary-complex assembly, ubiquitin loading, and 26 S engagement. From these, they infer that the rate-limiting step is not E2 discharge but the collision frequency between the lysine ε-amino group and the catalytic cysteine of the ubiquitin-charged E2. This observation would have been impossible with genetic methods, because they cannot deliver the temporal precision to disentangle conjugation from degradation. The same logic can be extended by chemical genetics to organisms that lack standard genetics. In filamentous fungi, a pomalidomide-based degrader was used to degrade the single essential E3 ligase. This showed that ubiquitin transfer continues for as long as the proteasome is intact but stops when the concentration of the ligase falls below threshold, providing direct, single-protein evidence for catalytic coupling between ligase abundance and substrate flux. Mass-spectrometry pipelines now link PROTAC treatment with diGly remnant profiling. This approach captures every ubiquitinated lysine across the proteome in a single run. Because the degrader is catalytic, only nanomolar concentrations are needed. This minimizes off-target solvent effects, allowing confident assignment of ubiquitin sites that are genuinely coupled to degradation rather than to stress-induced aggregation. The same dataset can be used to quantify chain linkage topology. It shows that substrates bound for complete proteolysis carry K48-linked tetra-ubiquitin whereas proteins tagged for regulatory purposes show mixed K11/K63 linkages, a distinction that was previously accessible only with antibody-based pulldowns that needed milligrams of starting material.
PROTACs as Tools in Chemical Biology and Proteomics
Chemical biology has embraced PROTACs as proximity-specific probes that report protein–protein interactions in cells. A pomalidomide-azide conjugate is attached to a photo-crosslinker and biotin reporter; upon UV irradiation, the new carbene inserts into any protein within 5 Å of the ternary complex, trapping transient interactors that would be lost in standard co-immunoprecipitation. Capture is followed by streptavidin enrichment and quantitative proteomics, providing an “interaction fingerprint” that discriminates direct ubiquitination partners from bystanders that merely co-sediment. Activity-based protein profiling (ABPP) has been integrated with PROTAC design to make degradation-only probes: the warhead is replaced by a covalent electrophile that labels the catalytic serine of a whole class of enzymes, while the pomalidomide moiety delivers the labelled proteins to the proteasome. The result is a chemical knockout that is limited to the active proteome fraction, and allows to ask whether the phenotype arises from the protein scaffold or from its enzymatic activity—a question that CRISPR cannot answer because it deletes both isoforms indiscriminately. Spatial proteomics leverages the same modular architecture. A pomalidomide-biotin conjugate is micro-injected into a defined cellular location (nucleolus, lamina, or synapse), and degradation is spatially restricted by adding a membrane-impermeant reducing agent that prevents the degrader from diffusing away. The ensuing loss-of-function is therefore spatially confined, and revealed that certain transcription factors are only required at the nuclear periphery and not throughout the nucleoplasm, a level of spatial resolution that is difficult to reach with global knock-down technologies. Finally, chemical proteomics pipelines deploy PROTAC libraries to map the “degradable kinome” or the “degradable deubiquitinome.” Each probe carries a fixed pomalidomide head and a variable kinase (or DUB) warhead; after treatment, lysates are analyzed by multiplexed mass spectrometry. Substrates that disappear across multiple chemotypes are deemed high-confidence degradable, whereas proteins that resist all analogues are classified as resistant and can guide future inhibitor design and reveal structural rules that govern degradability across entire protein families.
Table 2 Applications of PROTAC.
| Application | Probe Format | Read-out | Unique Advantage |
|---|
| Ubiquitin kinetics | Pomalidomide-fluorophore | Live-cell imaging | Minute-scale resolution |
| Cross-link interactome | Pomalidomide-azide-biotin | LC-MS/MS | Transient capture |
| Spatial degradation | Pomalidomide-biotin + redox gate | Immunofluorescence | Subcellular confinement |
| Degradable kinome | Pomalidomide-kinase warhead | Multiplex MS | Family-wide survey |
Industrial and Non-Pharmaceutical Applications
PROTAC chemistry is moving out of drug-discovery pipelines and into industrial biotechnology, where the same catalytic recruitment-ubiquitination-degradation principle is applied to regulate proteins in non-therapeutic contexts. Pomalidomide-derived CRBN recruiters are especially appealing in these applications because they are small, bioreactor-stable, and already qualified for synthesis at multi-kilogram scales. Agricultural formulators incorporate them in sprayable adjuvants to delete pest enzymes; synthetic-biology foundries link them to optogenetic switches to engineer self-editing microbial strains. The common denominator in all these applications is the need for a fast, irreversible way to target and delete a single protein without affecting the encoding gene (something CRISPR cannot do on a protein-by-protein basis).
PROTAC-Like Systems in Agricultural Biotechnology
Crop protection is shifting from non-specific toxophores to re-programmable protein eradication. The ideal weed-targeted degrader will bear a chloroacetamide warhead familiar to synthetic chemists, but rather than binding to the enzyme, it fastens the enzyme to a E3 ligase that the plant itself expresses. Ubiquitination takes place, and the protein of interest is removed before resistance-conferring mutations are selected. Since the same linker chemistry works with nucleotide, carbohydrate, or even peptide baits, "TF-PROTACs" have been designed to target the promoter binding domains of transcription factors which control cytochrome P450 expression. Eliminating these transcription factors switches off entire detoxification pathways and restores susceptibility to older herbicides without adding to the chemical burden. Field tests have also shown that nanomolar application rates are possible when degraders are formulated with lignin-based microparticles which protect the active ingredient from UV degradation until rain causes the formulation to release the active ingredient. Similar developments for insect control are harnessing the catalytic nature of the technology: A single foliar spray can degrade digestive proteases within caterpillars, while the same compound quickly mineralizes on leaf surfaces, safeguarding pollinator health. Regulatory bodies are also already asking for residue data to be framed around ubiquitin chain fragments, not the parent molecule, as it's understood that the pharmacologically active entity is the transient ternary complex, not the small molecule itself. PROTAC logic is therefore refocusing crop protection away from chemical warfare and towards precision protein editing, a strategy that should extend the commercial life of many existing active ingredients and lower environmental impact.
Protein Regulation Tools in Synthetic Biology and Diagnostics
Engineered cells that sense, compute and respond must be able to delete their own components on demand, a task for which PROTAC modularity is uniquely suited. The output phase of recent microbial design cycles has begun to insert a humanized E3 ligase under an arabinose promoter, followed by a matching degrader that displays a fluorophore quencher as its target ligand. Addition of the small molecule results in self-degradation of the reporter protein, an off-to-on fluorescence switch with a dynamic range that is impossible to match using transcriptional repression alone. Diagnostic developers have borrowed the same trick to create paper-based assays: a nasal swab lysate containing a viral protease is mixed with a PROTAC that displays a peptide epitope recognized by a colourimetric antibody. Protease cleavage separates the epitope from its E3-recruiting partner, thereby preventing ubiquitination and allowing accumulation of a colored immune complex within five minutes. Because the read-out is disappearance of ubiquitin conjugates rather than generation of a fluorescent signal, background interference from haem or mucus is negligible. In the longer term, CRISPR-free gene drives in yeast propose to couple degrader release to population density sensors; once a quorum is reached, a synthetic TF is eliminated, shutting down fertility genes and capping exponential growth. In each configuration the value proposition is identical—an abiotic small molecule that executes a post-translational deletion without altering DNA, thereby satisfying containment regulations that currently restrict genetically modified organisms.
Potential for Controlled Protein Recycling in Biomanufacturing
Industrial fermentations are commonly arrested when stress-responsive enzymes shunt carbon into futile repair cycles. Instead of the standard approaches of overexpressing chaperones or engineering expensive knockout strains, process engineers have recently begun to trial PROTAC-mediated clearance of the offending proteins. One degrader targeted against an acetate kinase, for example, was added to an E. coli culture in late log phase; the resulting decrease in acetate overflow channeled flux to the desired biopolymer and increased titre, without the genetic modification that would draw regulatory attention. Downstream, the same compound is quenched by addition of a high-affinity competitor, so that harvested biomass that will be used in food applications contains no active degrader. Similar logic is now being applied on a large scale in thousand-litre stainless reactors making therapeutic proteins. Host cell proteins that contaminate the product can be preemptively removed by a wash step that includes a broad-spectrum degrader cocktail; both the small molecules and the ubiquitinated contaminants are removed in subsequent depth filtration, simplifying chromatography and extending resin lifetime. Environmental officers are supportive of this approach because the effluent stream contains only short peptides rather than intact foreign proteins, reducing biological oxygen demand. In the future, continuous manufacturing suites will likely include inline sensors that meter degrader addition in response to real-time ubiquitin signals, thereby creating a self-correcting bioreactor that maintains proteostasis without human intervention. By turning protein degradation from a cellular accident into a dialled process parameter, PROTAC chemistry may tighten the link between metabolic modelling and industrial reality, leading to greener processes that use fewer resources to meet ever-stricter purity benchmarks.
The Role of Pomalidomide-Based Building Blocks
IMiD scaffolds have recently evolved from the clinical pomalidomide to universal chemical handles that anchor PROTAC programmes from academic labs to industry. Their bicyclic glutarimide imparts a pre-organised zinc-chelating motif that is recognised by CRBN orthologues in mammals, plants and insects, while the proximal aryl ring tolerates orthogonal linker attachment with minimal erosion of binding affinity. This synergy creates a plug-and-play module: labs stock one or two activated intermediates (C4-amine, C5-azide) and combine with any target ligand that presents the complementary handle. The operational outcome is a standardised degradation cassette that behaves predictably in biochemical, cellular and organismal assays, without the need to re-optimise E3 recognition for every new project.
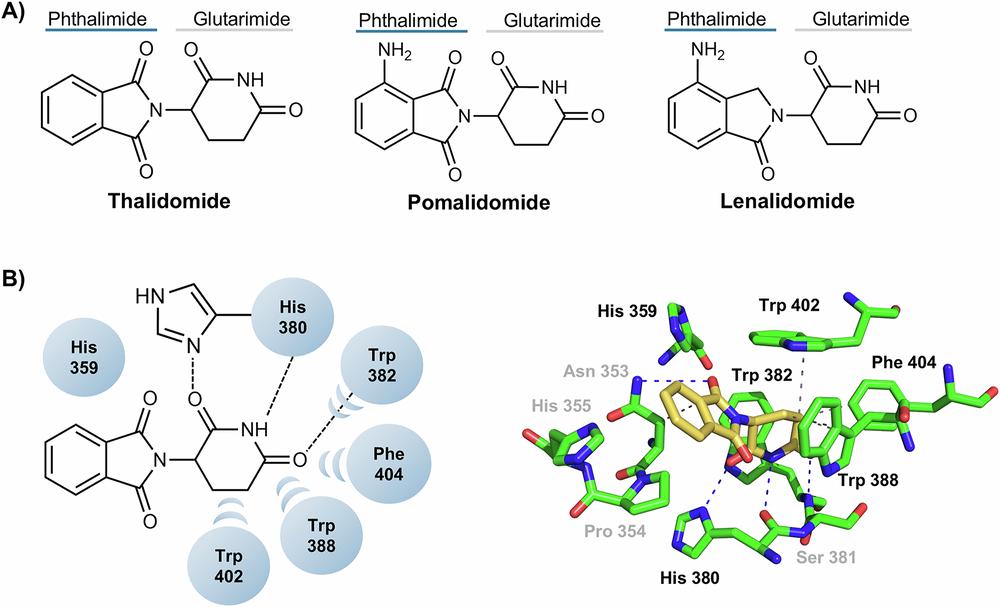 Fig. 2 Cereblon ligands.2,6
Fig. 2 Cereblon ligands.2,6
Advantages of CRBN Ligands for Research Flexibility
CRBN-targeting glutarimides provide additional experimental flexibility not afforded by the majority of other E3-targeting handles. Firstly, this binding site is a flat β-sheet groove on the thalidomide-binding domain, a surface that accommodates flat, spiro or even macrocyclic moieties without triggering the allosteric rearrangements that debilitate VHL ligands. As such, researchers can readily append nitrile, alkyne or azide functionalities directly onto the aryl ring without diminishing affinity, allowing for intracellular SuFEx or CuAAC ligation at a late stage—a strategy that would disrupt binding to MDM2 or IAP pockets. Secondly, CRBN is found in a wide range of vertebrates, insects and many filamentous fungi, meaning a single degrader can be translated from human cell culture lines to zebrafish embryos or tomato leaves without redesigning the E3 recruitment tag. Thirdly, the receptor is permissive of transient dissociation; as such, sub-stoichiometric doses are still able to induce quantifiable degradation, a property which is capitalized by synthetic chemists when residual small molecule cannot exceed parts-per-billion limits. Finally, CRBN ligands are orthogonal to native ubiquitin pathways, and so they neither induce compensatory E1 up-regulation nor activate heat-shock responses that can skew proteomic measurements. Together, these advantages make pomalidomide-based fragments chemically inert linkers that can be parked on any target ligand without interfering with endogenous signalling pathways.
Integration into Multi-Disciplinary Projects
The modular geometry of pomalidomide–linker conjugates has led to their use in applications well beyond oncology or even drug discovery. In plant biotechnology, a chloroacetamide herbicide was conjugated to pomalidomide using a short PEG bridge; spraying this degrader onto resistant weeds removed a key detoxifying glutathione transferase, restoring sensitivity to legacy herbicides without increasing environmental load. Materials scientists have tethered the same glutarimide to gold surfaces using a thiol linker, enabling chips to capture recombinant CRBN and subsequently ubiquitinate any prey protein delivered by microfluidics, thereby transforming solid-phase biosensors into self-resetting diagnostic platforms. In bio-manufacturing, formation of inclusion bodies during high-density fermentation was curtailed by feeding a pomalidomide-based degrader that selectively removed an aggregation-prone stress factor; the compound was later quenched with an excess of free thalidomide to ensure that downstream purification encountered no residual active molecule. Even astro-chemistry incubators have borrowed the scaffold, using photolabile pomalidomide derivatives to release amino-acid analogues under UV illumination and thereby modelling prebiotic peptide turnover on mineral surfaces. What unites these disparate applications is the recognition that pomalidomide functions as a generic CRBN-addressable tag whose biological liabilities in human therapy become irrelevant once the context is shifted to plants, materials or planetary chemistry. The molecule has thus metamorphosed from a restricted pharmaceutical into an open-source hardware component that any discipline can solder onto its own target of interest.
Our Academic and Industrial Support Portfolio
To support the expanding use of PROTAC chemistry beyond traditional drug discovery, we provide a comprehensive portfolio of research-grade materials, custom synthesis services, and collaborative development options tailored to both academic institutions and industrial R&D teams. Our solutions are designed to enable reliable experimentation, cross-disciplinary research, and scalable innovation, ensuring that PROTAC building blocks can be applied confidently in chemical biology, proteomics, synthetic biology, and emerging industrial applications.
Catalog of Research-Grade PROTAC Building Blocks
We maintain a curated catalog of high-purity PROTAC building blocks, including CRBN and VHL ligands, IMiD derivatives, validated linker libraries, and functional intermediates suitable for academic and industrial research. All compounds are supplied with full analytical characterization—HPLC, LC-MS, and NMR—ensuring reproducibility, traceability, and consistent performance across laboratories. These ready-to-use materials allow researchers to rapidly prototype degradation systems and explore new applications of targeted protein degradation.
Custom Synthesis and Collaboration for Specialized Applications
For projects requiring tailored molecular designs, we offer custom synthesis and collaborative development services for specialized PROTAC building blocks. Our chemistry team works closely with academic researchers and industrial partners to design application-specific ligands, linkers, and degradation tools, supporting exploratory research, platform development, and non-therapeutic protein regulation strategies. Each collaboration is guided by scientific feasibility, analytical rigor, and clear project milestones.
Advantage: Consistent Supply, Technical Support, and Co-Development Options
Our partners benefit from stable long-term supply, direct technical support from experienced chemists, and flexible co-development models that adapt to evolving research needs. By combining reliable manufacturing, in-depth PROTAC expertise, and transparent communication, we serve as a trusted partner helping academic and industrial teams advance protein degradation technologies efficiently and sustainably.
Work with a Trusted Partner for PROTAC Building Blocks
Explore our research-grade PROTAC building blocks or connect with our scientific team to discuss custom synthesis, collaborative projects, or specialized applications—we are ready to support your academic or industrial research with reliable chemistry, technical expertise, and flexible partnership models.
References
- Liu X, Wang A, Shi Y, et al. PROTACs in epigenetic cancer therapy: current status and future opportunities[J]. Molecules, 2023, 28(3): 1217. https://doi.org/10.3390/molecules28031217.
- Vicente A T S, Moura S P S P, Salvador J A R. Synthesis, biological evaluation and clinical trials of Cereblon-based PROTACs[J]. Communications Chemistry, 2025, 8(1): 218. https://doi.org/10.1038/s42004-025-01598-9.
- Cai J, Chen C, Wang J, et al. PROTAC: a revolutionary technology propelling small molecule drugs into the next golden age[J]. Frontiers in Oncology, 2025, 15: 1676414. https://doi.org/10.3389/fonc.2025.1676414.
- Liu Z, Hu M, Yang Y, et al. An overview of PROTACs: a promising drug discovery paradigm[J]. Molecular biomedicine, 2022, 3(1): 46. https://doi.org/10.1186/s43556-022-00112-0.
- Cardno A, Kennedy B, Lindon C. Cellular parameters shaping pathways of targeted protein degradation[J]. Communications Biology, 2025, 8(1): 691. https://doi.org/10.1038/s42003-025-08104-w.
- Distributed under Open Access license CC BY 4.0, without modification.

 Fig. 1 The structure and development process of protac: (a) The structure of PROTAC and the mechanism by which it targets protein degradation; (b) Major events in the history of PROTAC.1,6
Fig. 1 The structure and development process of protac: (a) The structure of PROTAC and the mechanism by which it targets protein degradation; (b) Major events in the history of PROTAC.1,6 Fig. 2 Cereblon ligands.2,6
Fig. 2 Cereblon ligands.2,6