The rapid rise of cereblon (CRBN)-based PROTACs has reshaped targeted protein degradation, offering researchers unprecedented control over intracellular protein levels. Yet as this technology expands, a critical challenge has come sharply into focus: undesired off-target degradation, particularly the unintended loss of zinc finger (ZF) transcription factors caused by IMiD-derived CRBN ligands such as pomalidomide. These off-target events not only distort biological readouts but also introduce potential safety concerns, making selectivity a defining requirement for next-generation degraders. This article provides a comprehensive overview of the mechanistic origins of CRBN-mediated ZF degradation, highlights proven strategies for reducing off-target effects, and outlines advanced experimental and design approaches that support the development of cleaner, more reliable PROTAC molecules.
Introduction — Understanding Off-Target Degradation
Off-target protein degradation has become one of the most critical challenges in the advancement of cereblon (CRBN)-based PROTAC technologies. Although PROTACs are engineered to induce selective and efficient elimination of disease-relevant proteins, the same molecular mechanisms that confer their potency can inadvertently trigger the degradation of unintended substrates. This phenomenon is especially relevant when using CRBN ligands derived from immunomodulatory drugs (IMiDs) such as pomalidomide, which are known to recruit a broad set of zinc finger (ZF) transcription factors. As a consequence, researchers must navigate the delicate balance between therapeutic efficacy and unintended biological liability.
From a drug development perspective, off-target degradation can lead to misleading mechanism-of-action data, reduced cellular viability unrelated to the intended target, and undesirable downstream transcriptional changes. In clinical settings, such off-target effects may contribute to adverse events, mirroring the well-documented pomalidomide side effects linked to ZF protein modulation. For teams optimizing next-generation degraders or building safer molecular glue profiles, understanding how these off-targets arise—and how to eliminate them—has become a strategic imperative.
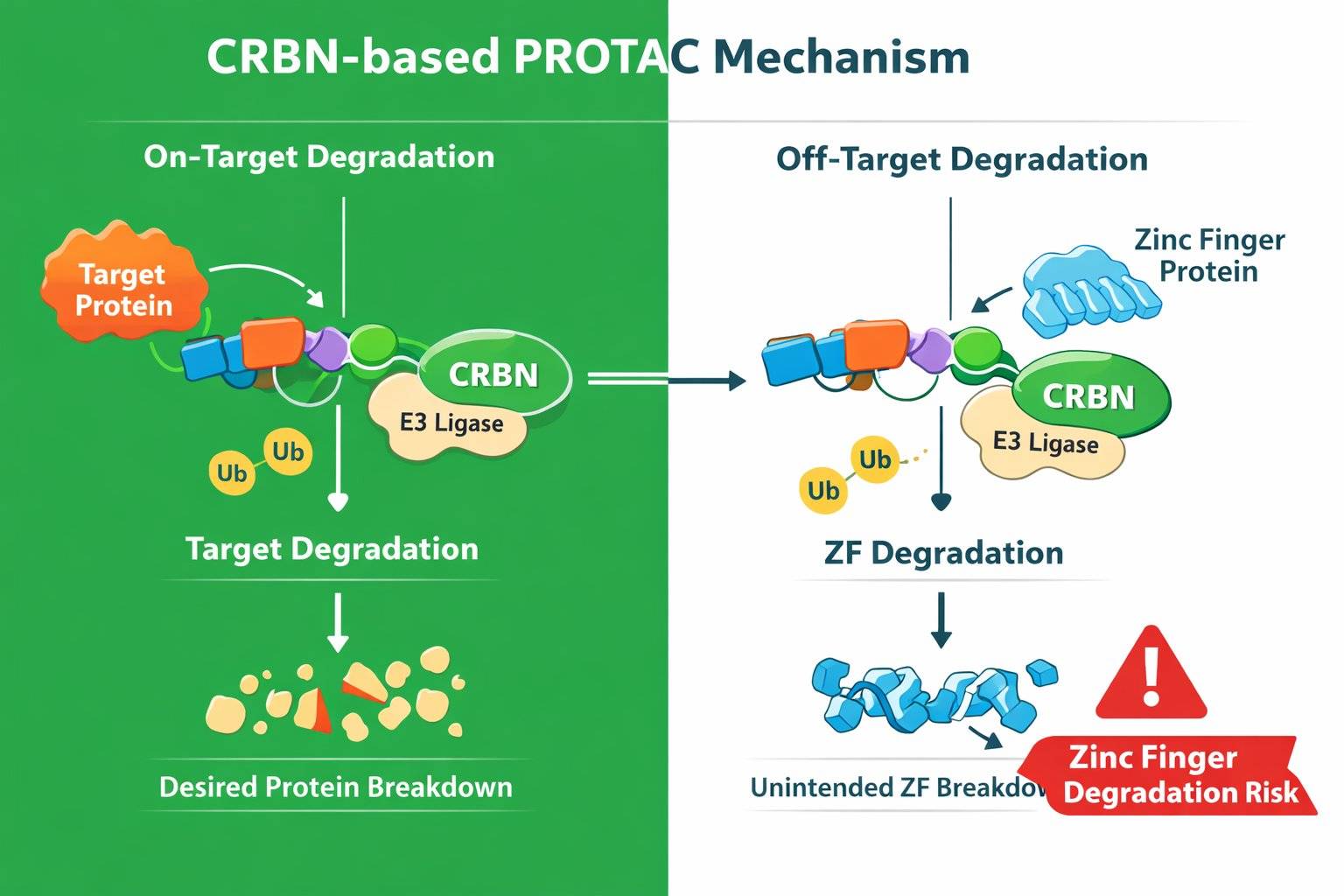 Fig 1. CRBN-based PROTAC mechanism
Fig 1. CRBN-based PROTAC mechanism
What Are Off-Targets and Why They Matter in PROTAC Research
In the context of targeted protein degradation, off-targets refer to proteins that are unintentionally ubiquitinated and degraded through the PROTAC-induced ternary complex. Unlike small-molecule inhibitors, whose off-target activity is often limited to transient binding, PROTAC-mediated degradation can create long-lasting and highly amplified biological consequences.
This distinction is especially important for CRBN-based degraders, where the E3 ligase ligand itself possesses inherent neosubstrate-recruiting properties. When a PROTAC inadvertently stabilizes a degrader-E3-off-target complex, even weak interactions can be converted into efficient ubiquitination events. The result: significant depletion of proteins never intended to be part of the therapeutic strategy.
| Feature / Mechanism | Intended Target Degradation | Off-Target ZF Degradation |
| Substrate Origin | Designed protein target | Unintended ZF transcription factors |
| Binding Driver | High-affinity ternary complex | Weak interactions amplified by IMiD-induced neosubstrate recruitment |
| Selectivity | High (engineered by PROTAC design) | Low (CRBN pocket permissiveness) |
| Biological Impact | Desired therapeutic effect | Dysregulated transcription, adverse phenotypes |
| Risk Level | Controlled | High risk for assay noise & toxicity |
The Case of Zinc Finger Protein Degradation
Among all off-target liabilities associated with CRBN, zinc finger proteins represent the most sensitive and widely affected class. Many ZF transcription factors contain structural motifs that mimic known IMiD-bound neosubstrates, allowing them to be mistakenly recruited to CRBN when pomalidomide-based ligands are used. The degradation of these ZF proteins can disrupt gene regulation networks, impair cell differentiation, and introduce phenotypic noise into experimental systems.
For PROTAC developers, this poses substantial risks: inaccurate interpretation of biological assays, diminished selectivity profiles, and potential safety concerns if the degrader moves into translational studies. Consequently, mitigating CRBN off-target degradation—particularly of ZF proteins—has become a foundational objective for designing next-generation, high-specificity PROTACs.
Mechanistic Basis of CRBN-Mediated Off-Targeting
Understanding why CRBN-based PROTACs induce unintended degradation requires a close look at the interplay between the E3 ligase, its molecular glue-like ligands, and structurally susceptible neosubstrates. Unlike other E3 ligases commonly used in targeted protein degradation, CRBN possesses a unique binding pocket that naturally accommodates a wide variety of zinc finger (ZF) domains. When pomalidomide or related IMiDs are incorporated into PROTAC designs, this pocket becomes even more permissive, dramatically increasing the likelihood of unintended protein recruitment.
This mechanistic vulnerability—rooted in CRBN's intrinsic substrate preferences—is the foundation of most off-target degradation events observed with CRBN-directed degraders. By dissecting this molecular recognition process, researchers can strategically redesign CRBN ligands to retain target engagement while suppressing unwanted ZF binding.
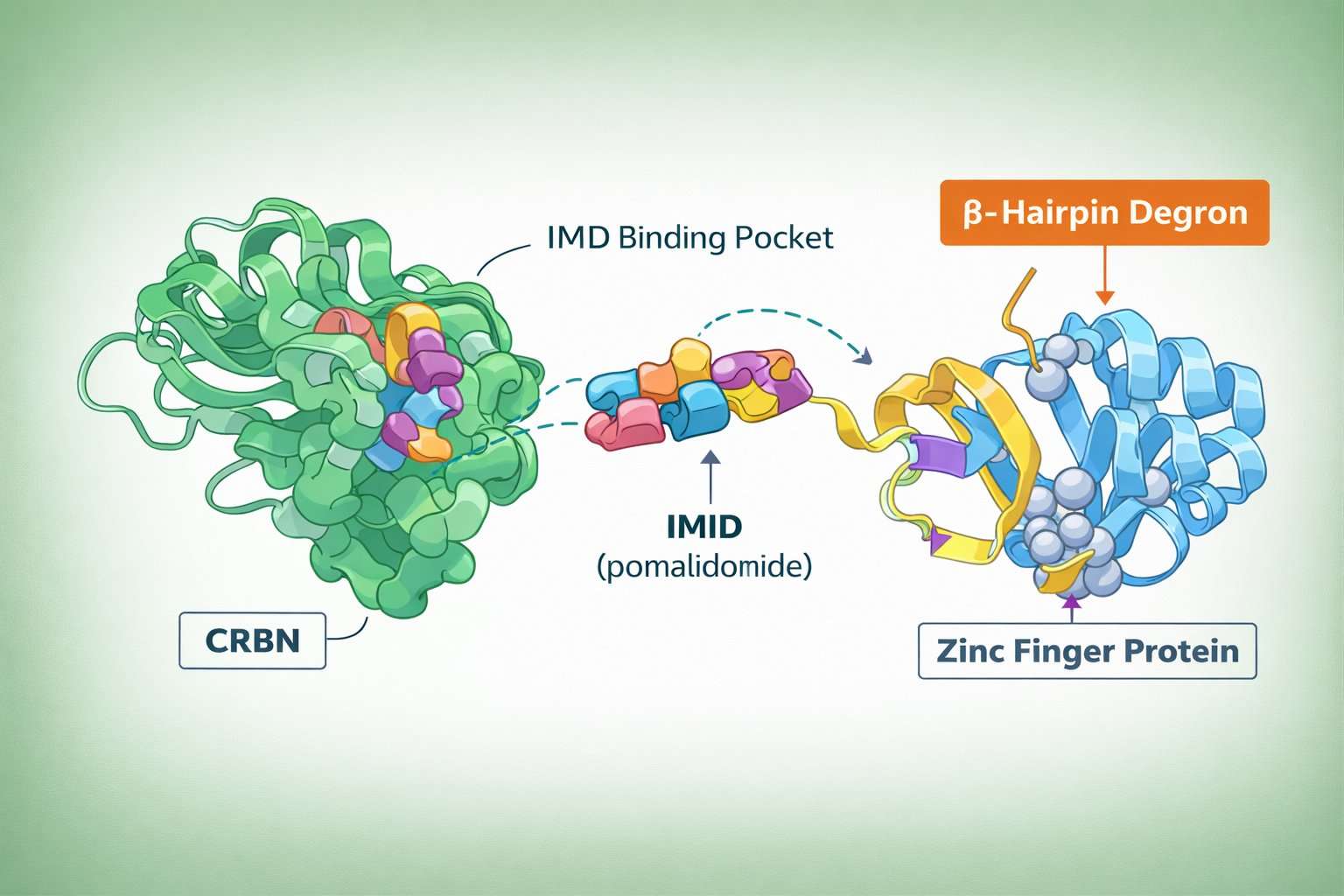 Fig 2. A scientific molecular illustration of CRBN, IMiD (pomalidomide), and zinc finger protein interaction
Fig 2. A scientific molecular illustration of CRBN, IMiD (pomalidomide), and zinc finger protein interaction
CRBN-IMiD-ZF Protein Interaction Model
The interaction between CRBN, IMiD-derived ligands, and zinc finger proteins is now well understood due to extensive crystallographic and proteomics studies. Classic IMiDs such as pomalidomide and lenalidomide bind to CRBN's thalidomide-binding domain and enable it to recruit neosubstrates that would not normally interact with this ligase.
In this model:
- IMiD binding reshapes the CRBN surface, creating a new interface capable of engaging Cys2His2 ZF motifs.
- ZF proteins containing a "degron-like" β-hairpin are efficiently captured and ubiquitinated.
- Even weak or transient interactions become degradation-competent, because the PROTAC ternary complex effectively stabilizes the ligase-substrate interface.
This explains why ZF transcription factors are disproportionately affected when CRBN ligands are used: they possess the structural and electrostatic features that align with the CRBN-IMiD pocket, making them highly susceptible to unintended degradation.
Structural Reasons for Unintended Protein Recruitment
CRBN's susceptibility to off-target recruitment is rooted in a combination of structural features:
- A highly adaptable substrate-binding pocket: The CRBN pocket tolerates diverse chemical substituents and can accommodate secondary interactions introduced by PROTAC linker positioning.
- Pomalidomide's exposed glutarimide and phthalimide moieties: These functional groups interact with conserved residues within CRBN, while also presenting conformations favorable for contacting ZF proteins.
- ZF domains featuring solvent-exposed degrons: Many ZF proteins naturally present their β-hairpin surfaces, making them readily captured upon IMiD binding.
- Ternary complex stabilization increases recognition efficiency: Even substrates with marginal affinity can be pulled into a productive degradation complex once both ends of the PROTAC are engaged.
Together, these structural elements create an environment where a designed PROTAC may unintentionally amplify CRBN's innate ZF-binding tendencies. This molecular understanding has become the basis for modern strategies focused on reducing ZF off-target degradation—primarily through deliberate modifications to the pomalidomide scaffold and through rational design of CRBN ligands with minimized neosubstrate recruitment potential.
Strategies to Reduce Zinc Finger Degradation
Minimizing off-target zinc finger (ZF) protein degradation has become a central objective in the development of next-generation CRBN-based PROTACs. Modern degrader design now focuses not only on potency and target engagement but also on the judicious engineering of cereblon ligands to suppress unintended neosubstrate recruitment. By modifying specific chemical features, redesigning ligand-protein interfaces, and leveraging computational prediction tools, researchers can achieve a significantly cleaner degradation profile without sacrificing efficacy.
Chemical Modification at the Pomalidomide C5 Site
One of the most validated approaches for reducing ZF off-target degradation is structural modification of pomalidomide at the C5 position. Numerous studies have shown that the C4-C5 region plays a critical role in neosubstrate recognition, particularly in interactions involving ZF degron motifs. Key strategies include:
- Introducing steric bulk at C5: Added steric hindrance can block ZF degron engagement while retaining CRBN binding affinity. This shifts the degradation selectivity toward the intended target.
- Incorporating electron-donating or withdrawing substituents: Fine-tuning electronic properties can alter the binding surface, making it less compatible with ZF recognition.
- Expanding or altering ring substituents for linker attachment: C5-modified exit vectors often maintain full ternary complex formation with the desired target while dramatically reducing ZF protein recruitment.
Empirically, many next-gen pomalidomide analogs with C5 modifications demonstrate 10-50× lower ZF degradation, marking this as a cornerstone technique for selectivity optimization.
Designing Selective Ligands with Minimal ZF Affinity
Beyond C5 modification, researchers now explore completely reengineered CRBN ligands that naturally exhibit low ZF affinity. These selective ligands are designed based on:
- Structure-guided disruption of known ZF degron-contact residues
- Reduced complementarity to ZF β-hairpin motifs
- Optimized physicochemical properties that favor target-specific ternary complexes
Examples include novel CRBN ligands lacking the classical IMiD glutarimide orientation, or possessing altered amide linkers, both of which weaken undesirable neosubstrate binding. These ligands often preserve strong target degradation potency while reducing signature ZF protein loss seen in global proteomics assays.
Computational Prediction of Neosubstrate Binding
Advances in computational modeling have introduced predictive frameworks that help researchers identify ZF off-target risks before synthesizing new PROTACs. Common in silico methodologies include:
- Molecular docking and protein-ligand interaction scoring: Used to estimate whether a designed CRBN ligand favors ZF-like interfaces.
- Ternary complex simulation and stability analysis: Predicts whether CRBN, the ligand, and potential ZF proteins form energetically favorable complexes.
- Machine learning models trained on proteomics datasets: These tools predict degron-likeness across thousands of ZF proteins, guiding medicinal chemists toward safer ligand designs.
With these computational insights, chemical modifications can be prioritized to reduce off-target activity, enhance selectivity, and shorten development timelines.
| Strategy | Mechanism | Expected Benefit | Limitations |
| C5 modification on pomalidomide | Introduces steric/electronic changes to block ZF degron binding | 10-50× reduction in ZF degradation | Must preserve CRBN affinity |
| Selective CRBN ligands | Re-engineered IMiD scaffold to inherently avoid ZF recruitment | Near-complete elimination of ZF off-targets | Higher synthetic complexity |
| Computational neosubstrate prediction | Predict ZF-binding risks pre-synthesis | Reduces chemistry cycles; early risk control | Requires dataset/model accuracy |
| Proteomics screening | Measures global degradation patterns | Definitive verification of selectivity | Higher cost & time |
Experimental Approaches and Case Studies
Designing CRBN-based PROTACs with minimal zinc finger (ZF) off-target degradation requires not only rational chemistry but also robust experimental validation. Modern labs now rely on integrated proteomics, imaging, and case-study benchmarking to quantify off-target risks and compare ligand designs. These data-driven strategies ensure that structural modifications translate into meaningful biological improvements.
Proteomics-Based Off-Target Screening
Proteomics has become the gold standard for assessing unintended substrate degradation. By measuring global protein abundance after PROTAC treatment, researchers can systematically identify ZF proteins—and other neosubstrates—that are inadvertently recruited by CRBN. Key approaches include:
- TMT-based quantitative proteomics: Allows multiplexed comparison of many ligand variants, providing high-resolution insight into differential ZF degradation.
- Time-course degradation profiling: Differentiates early, direct CRBN-mediated off-targets from downstream transcriptional or signaling effects.
- Degron-mapping and motif enrichment analysis: Identifies ZF domains with high susceptibility to CRBN recruitment and helps refine ligand design strategies.
These assays have consistently demonstrated that small modifications at the CRBN ligand's C5 or C4 positions can drastically reduce ZF degradation while maintaining on-target activity.
High-Throughput Image Analysis of Degradation Profiles
While proteomics provides depth, high-throughput imaging offers speed and phenotypic clarity. Automated microscopy combined with AI-driven image analysis allows rapid quantification of target and off-target degradation across large cell panels. Typical readouts include:
- Fluorescence-tagged protein depletion kinetics: Tracks degradation efficiency in real time, enabling direct comparison of PROTAC variants.
- Cell viability and morphological assessments: Helps determine whether observed cytotoxicity stems from the intended target or from broader ZF transcriptional dysregulation.
- Subcellular localization studies: Reveal whether structural modifications alter PROTAC trafficking or engagement with E3 ligase machinery.
This approach is especially useful during early-stage analog screening, where rapid feedback accelerates iterative medicinal chemistry cycles.
Successful Low-ZF-Affinity PROTAC Examples
Several published and proprietary case studies highlight the success of structural engineering in suppressing ZF off-target degradation:
- C5-modified pomalidomide PROTACs: These exhibit dramatically reduced degradation of IKZF1, ZNF653, and other known IMiD-sensitive ZF proteins—while maintaining potent degradation of oncogenic and epigenetic targets.
- Novel CRBN ligands engineered for reduced ZF recruitment: Some analogs demonstrate near-complete elimination of ZF off-target activity, achieving unprecedented selectivity profiles suitable for translational development.
- Hybrid E3 ligase ligands with altered exit vectors: These compounds maintain CRBN engagement but redirect steric and electrostatic surfaces to avoid ZF degron mimicry.
Case studies consistently demonstrate that rational design, supported by empirical and computational validation, can reduce off-target ZF degradation by an order of magnitude or more—without sacrificing efficacy against the intended disease-relevant protein.
Our Solutions for Selective PROTAC Development
As the industry shifts toward safer, more selective targeted protein degraders, researchers increasingly require CRBN ligands and analytical support that minimize off-target zinc finger (ZF) degradation without compromising potency. Our platform integrates medicinal chemistry, structural biology, and advanced profiling technologies to deliver highly selective pomalidomide analogs and data-driven PROTAC optimization services. These solutions help teams accelerate development, reduce risk, and generate cleaner mechanism-of-action data.
Low Off-Target Pomalidomide Analogs with Verified Selectivity
We offer a portfolio of next-generation CRBN ligands engineered specifically to suppress ZF neosubstrate recruitment. Each analog is designed using structure-driven modifications at key positions—most notably the C5 site—to reduce affinity toward IMiD-sensitive ZF domains while preserving strong CRBN binding. Key advantages include:
- Substantially reduced ZF protein degradation in proteomics and imaging assays
- High stability in ternary complex formation with intended targets
- Optimized exit vectors compatible with diverse linker chemistries
- Improved safety profiles for translational research and preclinical studies
These ligands serve as ideal starting points for PROTAC programs seeking improved selectivity and lower off-target liabilities.
Advanced Off-Target Profiling and Analytical Support
To support rational design and regulatory readiness, we provide a suite of analytical services that help developers quantify and minimize off-target activity:
- Global proteomics for off-target identification: Comprehensive analysis of ZF and other CRBN-sensitive proteins at varying doses and time points.
- Degradation kinetics and half-life measurements: High-resolution evaluation of both intended and unintended targets.
- Computational modeling of neosubstrate risk: Predictive analysis for ligand optimization prior to synthesis, reducing trial-and-error cycles.
- Comparative selectivity benchmarking: Enables clear comparison across multiple PROTAC candidates or CRBN ligand scaffolds.
These capabilities ensure robust data packages for internal development decisions, publications, and regulatory documentation.
Advantage: Cleaner Mechanisms, Higher Reliability, and Regulatory Compliance
PROTACs designed with our selective CRBN ligands demonstrate:
- Cleaner biological readouts: Reduced noise from ZF-driven transcriptional disruptions leads to more accurate interpretation of target biology.
- Lower safety risks: Minimizing off-target ZF degradation aligns with regulatory expectations for mechanism-of-action clarity and toxicology consistency.
- Higher development efficiency: Early removal of off-target liabilities prevents downstream failure, saving time and resources.
By integrating selectivity-optimized chemistry with rigorous analytical validation, we help research teams advance PROTAC programs with greater confidence and superior scientific quality.
Get Expert Support for CRBN Selectivity and PROTAC Optimization
Achieving high selectivity in CRBN-based PROTACs is no longer optional—it is a fundamental requirement for reliable mechanism-of-action studies, safer preclinical profiles, and successful downstream development. Whether you are troubleshooting unexpected zinc finger degradation or proactively optimizing your degrader chemistry, our team offers end-to-end scientific support designed to help you overcome off-target challenges with confidence. Ready to advance your PROTAC program with higher selectivity and fewer off-target risks? Contact our technical team today to schedule a consultation or request pricing for our selective CRBN ligands.

 Fig 1. CRBN-based PROTAC mechanism
Fig 1. CRBN-based PROTAC mechanism Fig 2. A scientific molecular illustration of CRBN, IMiD (pomalidomide), and zinc finger protein interaction
Fig 2. A scientific molecular illustration of CRBN, IMiD (pomalidomide), and zinc finger protein interaction