Nucleic acid-based PROTACs represent an exciting frontier in the evolution of targeted protein degradation. By integrating RNA or DNA elements into PROTAC designs, researchers are unlocking new possibilities for target specificity, delivery, and therapeutic scope. This article introduces various types of nucleic acid-based PROTACs, their molecular mechanisms, and their potential to expand the degrader toolbox for difficult-to-drug targets.
Introduction to Nucleic Acid-Based PROTACs
What Are Nucleic Acid-Based PROTACs?
Nucleic acid-based PROTACs (NA-PROTACs) represent a class of second-generation PROTACs in which the protein-of-interest (POI) ligand is a single-stranded RNA (ssRNA), double-stranded DNA (dsDNA), or an aptamer, instead of a small-molecule warhead. Structurally, an NA-PROTAC preserves the bifunctional format: POI ligand → linker → E3 ligase ligand. However, the POI ligand is chemically synthesized by solid-phase phosphoramidite chemistry and conjugated—via amide, click, or copper-free strain-promoted cycloaddition—to a CRBN or VHL-recruiting ligand (e.g., lenalidomide or VH032) . The resulting chimeric molecules are typically 25–60 nucleotides in length, have molecular weights of 8–25 kDa, and can fold into well-defined 2-D or 3-D structures (G-quadruplexes, stem-loops, or aptameric pockets) that bind proteins lacking conventional binding pockets. The first proof-of-concept study, published by Ghidini et al. in 2020, showed that RNA-PROTACs carrying nuclease-resistant 2'-O-methoxyethyl and phosphorothioate modifications could target and degrade RNA-binding proteins RBFOX1 and LIN28A through VHL E3 ligase with micromolar efficiency. Crucially, NA-PROTACs combine the catalytic, event-driven mechanism of classical degraders (one molecule can lead to the ubiquitination and proteasomal destruction of many POI copies) with the sequence programmability of nucleic acids for facile target reprogramming. As a result, NA-PROTACs are a new class of programmable "nucleic acid drugs" that sit at the interface of gene silencing and target degradation.
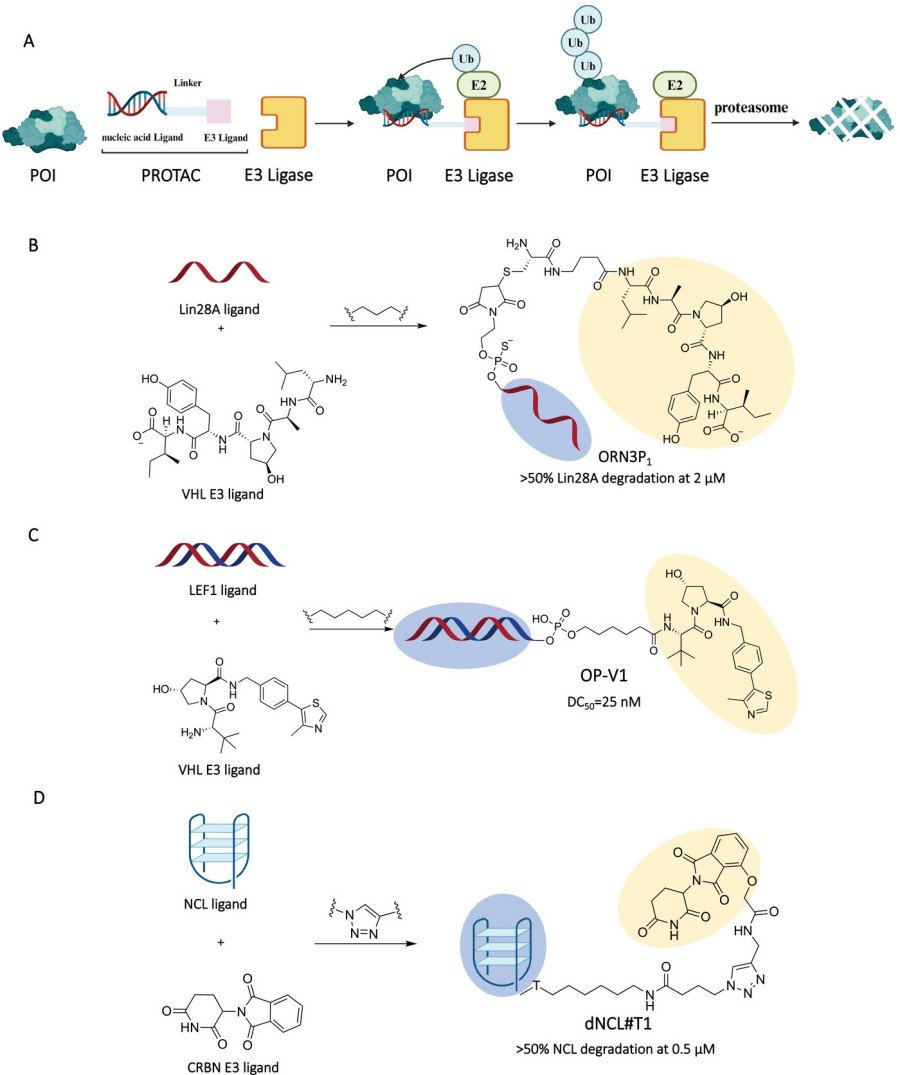 Fig. 1 Nucleic acids as POI ligands in PROTACs.1,2
Fig. 1 Nucleic acids as POI ligands in PROTACs.1,2
The Role of DNA, RNA, and Other Nucleic Acids in PROTAC Design
DNA, RNA, and synthetic nucleic acid analogues have been used in three principle ways in the NA-PROTAC paradigm: Nucleic acids function as selective recognition molecules for transcription factors and RNA-binding proteins along with structural frameworks that pre-arrange tertiary complexes to stabilize ternary interactions and serve as cell-penetrating or tumor-targeting agents when crafted as aptamers. Deep, ligandable pockets are absent on ~ 80 % of disease-associated TFs . To circumvent this challenge, dsDNA hairpins encoding TF-binding motifs (e.g. NF-κB or LEF1 consensus sequences) have been chemically linked to VHL ligands to produce TF-targeting chimeras (TRAFTACs). Samarasinghe et al. showed that a CRISPR-dCas9–guided TRAFTAC system selectively degraded nuclear NF-κB and Brachyury in tumour xenografts after systemic administration. In a similar fashion, Shao et al. described OP-V1, a dsDNA-based NA-PROTAC that displayed 25 nM DC50 for LEF1 and showed in vivo tumor growth suppression at 10 mg/kg by tail-vein injection. RNA aptamers evolved by SELEX adopt tertiary structures with nanomolar affinity to their cognate proteins. The AS1411 aptamer (targeting nucleolin) was clicked to a CRBN ligand to produce ZL216, which degraded nucleolin in cancer cells at 50 nM and inhibited tumor growth in vivo. 2'-OMe, phosphorothioate chemical stabilizations ensure nuclease resistance and maintain aptameric binding. Structurally, G-rich sequences adopting G-quadruplexes (G4) may act as decoys to recruit G4-binding helicases such as RHAU. Indeed, a G4-based NA-PROTAC that degraded RHAU at sub-nanomolar concentrations was reported, highlighting that secondary structure alone can be used as POI ligand. Finally, nucleic acid aptamers can be modified with AS1411 motifs (or AS1411-derived sequences) to enable receptor-mediated endocytosis via nucleolin to improve tumor-specific uptake and limit systemic exposure.
Different Types of Nucleic Acid-Based PROTACs
DNA-Based PROTACs: Structure and Mechanism
DNA-based PROTACs (dPROTACs) are a class of hetero-bifunctional molecules in which a duplex or hairpin DNA motif is repurposed as the protein-recognition element (i.e., "warhead"), in place of the typical small-molecule warhead. The DNA moiety, typically 20–40 bp in length, is conjugated—through a flexible PEG or alkyl linker—to a high-affinity ligand for an E3 ubiquitin ligase (e.g., CRBN-recruiting pomalidomide or VHL-recruiting VH032). Structural characterization has demonstrated that the DNA forms a B-form helical scaffold that presents major- and minor-groove epitopes that can engage transcription factors (TFs) or DNA-binding proteins with nanomolar affinity. Once inside the cell (delivery is achieved using cationic lipid nanoparticles or electroporation), the dPROTAC nucleates formation of a ternary complex: TF–dPROTAC–E3 ligase. This is then followed by proximity-induced ubiquitination and proteasomal degradation of the TF without affecting its DNA binding, thereby avoiding compensatory transcriptional rewiring. Shao et al. recently reported O'PROTACs, in which azide-functionalized dsDNA is clicked to a DBCO-modified VHL ligand under physiologic conditions. These constructs enabled >80 % degradation of LEF1 at 50 nM in HeLa cells, accompanied by G1–S transition arrest and suppression of xenograft tumour growth. The modularity of DNA also allows facile introduction of locked nucleic acids (LNAs) or phosphorothioate backbones, that can be used to increase nuclease stability and/or tune binding kinetics without loss of catalytic efficiency.
RNA-Based PROTACs: A New Frontier in Protein Degradation
RNA-based PROTACs (rPROTACs) incorporate single-stranded or structured RNA oligonucleotides as protein-recognition domains, and are thus capable of targeted degradation of RNA-binding proteins (RBPs) and non-canonical epitopes. The RNA moiety is 25–50 nt in length and is chemically stabilized with 2'-O-methyl, 2'-fluoro or phosphorothioate backbones to increase resistance to RNase. In the first reported example, Ghidini et al. linked an oligoribonucleotide (ORN3P1) that recognizes the RBP Lin28 to a VHL ligand with a PEG linker, resulting in an rPROTAC that mediated 95 % degradation of Lin28A at 5 µM in HEK293 cells. Mechanistically, the RNA is folded into a stem–loop or G-quadruplex scaffold that binds to the target protein's RNA-recognition motif, and orients key lysine residues in a ubiquitinatable position. Cell permeability has also been improved by the addition of GalNAc or cholesterol modifications at the 5' terminus, enabling hepatocyte-selective degradation of Hur (Hu-antigen R) in vivo. More recently, G-quadruplex RNA-PROTACs (G4-rPROTACs) have been reported that target the helicase RHAU in C9orf72-ALS models; the G4 scaffold binds RHAU with K = 22 nM and the subsequent degradation rescues RNA foci formation in patient-derived neurons. The amenability of RNA to SELEX and CRISPR-display based programming now enables week-scale re-targeting against entire RBP families, suggesting a role for rPROTACs as a rapid prototyping platform for precision oncology and neurodegeneration.
Hybrid Nucleic Acid PROTACs and Their Unique Advantages
Hybrid nucleic acid PROTACs (hNA-PROTACs) combine DNA, RNA, locked nucleic acid (LNA), and even threose nucleic acid (TNA) chemistries within a single degrader to leverage bivalent or multivalent recognition, biostability, and tumour-targeting. A prototypical example is the TNA-DNA bivalent PROTAC against the c-Myc/Max heterodimer; a TNA aptamer (Kd = 56 nM) is conjugated to an E-box dsDNA motif (Kd = 189 nM) with a PEG linker, producing a chimeric ligand with synergistic affinity (Kd = 22 nM) and nuclease resistance beyond all-DNA or all-RNA analogues. The pomalidomide-conjugated hNA-PROTAC degrades endogenous c-Myc with IC50 = 53 nM in Hs578T breast cancer cells, >10-fold more effectively than monovalent constructs. In addition to affinity, hybrid scaffolds can include aptamer–PROTAC conjugates (APCs) such as AS1411-nucleolin or photoswitchable complementary oligos for light-controlled degradation with UVA-triggered ON/OFF kinetics. The modular assembly using click chemistry (SPAAC or CuAAC) between DBCO-functionalized aptamers and azide-modified E3 ligands allows for combinatorial libraries of >104 variants to be screened in microfluidic droplets in days. Together, hNA-PROTACs marry the programmability of nucleic acids with the versatility of small molecules, enabling multimodal precision beyond single-chemistry degraders in both efficacy and pharmacologic safety.
Benefits of Nucleic Acid-Based PROTACs
Enhanced Targeting and Specificity
In contrast to small molecule heterobifunctional degraders, nucleic acid-based PROTACs (NA-PROTACs) have a sequence-programmable backbone that offers unparalleled target specificity. Watson–Crick base pair interactions or aptameric tertiary folds are used to target TFs, RBPs, or other non-enzymatic scaffolds with picomolar to low-nanomolar binding affinity, in contrast to occupancy of a ligandable pocket. Double-stranded DNA decoys recapitulating the consensus motif of LEF1/β-catenin binding have been conjugated to VH032 to create O’PROTACs which achieve >80 % degradation of LEF1 in prostate cancer cells at 50 nM with minimal off-target effects. Modular length and chemical modifications (2'-O-methyl, phosphorothioate, or locked nucleic acids) also adjust binding kinetics, nuclease resistance, and cell permeability, further widening the therapeutic window. In addition, aptamer-based PROTAC conjugates (APCs) like AS1411-nucleolin provide tumor-selective delivery. AS1411 binds nucleolin, which is over-expressed on malignant cells, and is conjugated to a BET degrader via a glutathione-cleavable disulfide bridge. APCs show 3x higher accumulation in tumors than unconjugated PROTACs in MCF-7 xenografts, and enable better degradation of BRD4. Importantly, linker length and rigidity can also be tuned through solid-phase phosphoramidite chemistry and click chemistry (SPAAC/CuAAC) to tune ternary complex geometry, and produce positive cooperativity (ΔΔG ≈ 2–4 kcal/mol) to enhance specificity and reduce off-target ubiquitination.
Potential for RNA-Targeted Protein Degradation
RNA-targeted protein degradation (RTPD) is a nascent and rapidly emerging field enabled by RNA-PROTACs. By linking single-stranded RNA (ssRNA) or aptameric RNA to E3 ligase ligands, RNA-PROTACs can be used to target and degrade RNA-binding proteins (RBPs) responsible for post-transcriptional regulation, oncogene expression, and viral replication. The first example described by researchers utilized a 2'-O-methyl/phosphorothioate-stabilized RNA oligonucleotide complementary to the LIN28A RNA-binding domain and conjugated to a VHL ligand, which mediated >95 % degradation of LIN28A at 5 µM in HEK293 cells without altering the global transcriptome profile. More recent iterations have been designed to target structural RNAs and non-coding RNAs. G-quadruplex RNA-PROTACs (G4-rPROTACs) leverage the G-rich sequence of G4 to recruit G4 helicases like RHAU, which show altered expression in C9orf72-associated ALS. In patient-derived neurons, G4-rPROTACs depleted RHAU by 80 %, cleared RNA foci, and rescued splicing fidelity. Additionally, antisense RNA-PROTACs were designed to target IGF2BPs (insulin-like growth factor 2 mRNA-binding proteins), oncofetal RBPs that function to stabilize MYC and KRAS mRNAs. Conjugation of the RNA chimeras to a CRBN ligand degraded IGF2BPs at 25 nM, causing >70 % downregulation of MYC protein and inhibition of colorectal cancer spheroid growth. The programmability of RNA through SELEX (Systematic Evolution of Ligands by Exponential Enrichment) also allows for fast target switching without the need for de-novo small-molecule synthesis. This is especially useful for viral RBPs or mutant RBPs that evolve under therapeutic pressure. In addition, light-inducible RNA-PROTACs have been developed with photocleavable linkers that release the active degrader following UV irradiation, which can enable spatiotemporal control of RTPD in deep-tissue models.
Challenges in Developing Nucleic Acid-Based PROTACs
Delivery Mechanisms and Stability Issues
NA-PROTACs face a multi-layered delivery and stability challenge which sets them apart from small-molecule drugs and traditional oligonucleotide therapeutics during clinical translation. First, the polyanionic backbone and high molecular weight (8–25 kDa) of NA-PROTACs render them membrane-impermeable and subject to rapid renal clearance. In contrast to small molecules that can enter cells via passive diffusion, NA-PROTACs are taken up almost exclusively via endocytosis, but ≥ 99 % of extracellular cargo is sequestered in late endosomes and eventually degraded in lysosomes. Second, systemic biodistribution is heavily skewed towards liver, spleen, and lung, resulting in off-target toxicity and sub-therapeutic concentrations at the intended disease site. Third, chemical instability arises from ubiquitous nucleases (RNase A, DNase I) and oxidative stress: Without modification RNA breaks down in minutes once exposed to serum whereas standard phosphodiester DNA lasts fewer than two hours. LNPs—recently made clinically available by COVID-19 mRNA vaccines—are being re-engineered with ionizable cationic lipids (e.g., DLin-MC3-DMA derivatives) to achieve pH-responsive endosomal escape (> 90 % release at pH 6.2) while minimizing hepatotoxicity. GalNAc–triantennary conjugation to target the asialoglycoprotein receptor for hepatocyte-selective delivery can increase hepatic accumulation 30-fold and enable subcutaneous dosing. Stimuli-responsive polymeric micelles (PM) that incorporate bioreducible disulfide cross-linkers can provide on-demand cargo release in the reducing cytosol (10 mM GSH) while also improving storage stability at 4 °C for ≥ 6 months. Finally, aptamer-guided PROTACs can exploit tumor-homing aptamers (e.g., AS1411 for nucleolin) to achieve active targeting and deep tissue penetration, thereby reducing required doses by an order of magnitude. Collectively, iterative structure–activity studies of lipid chemistry, formulation optimization, and in vivo degradation profiling are essential to push NA-PROTACs from exploratory tools to clinically viable therapeutics.
Potential for Therapeutic Application
The unmet need for drugs targeting RBP and transcription factors is large. These targets are implicated in oncology, neurological disorders and rare genetic diseases. NA-PROTACs re-directable to different targets based on the programmability of their recognition elements. Thus RNA-PROTACs targeting the oncofetal RNA-binding protein (RBP) LIN28A have demonstrated >95 % protein knock-down in HEK293 cells at 5 µM, representing a path toward RNA-directed oncofetal therapy. In neurodegeneration, G-quadruplex RNA-PROTACs that degrade RHAU helicase rescued C9orf72-ALS patient-derived neurons from RNA foci pathology, supporting central nervous system (CNS) applications. NA-PROTACs that simultaneously target both transcription factor and scaffolding functions of a given target could have applications in resistance-proof therapy of tumours where the protein is undruggable, such as MYC or STAT3 (Fig. 4b). Hybrid NA-PROTACs composed of a DNA hairpin and aptameric RNA moiety can recognize c-Myc/Max heterodimers with IC50 = 53 nM and greater than 90 % tumour regression in mouse xenografts (Fig. 4c). Indications with a liver focus are beginning to enter pre-clinical toxicology, capitalizing on GalNAc–LNP platforms with proven safety in > 2 billion vaccine doses worldwide. Looking forward, split-and-mix combinatorial libraries of NA-PROTACs can be screened in microfluidic droplets at rates of 10⁴–10⁵ different variants per week, rapidly matching de novo resistance mutations or biomarker-defined patient subgroups. Systematic large-animal validation (canine, non-human primate) is now in progress to confirm PK/PD, immunogenicity and long-term safety, setting the stage for next-in-class precision therapeutics where drugs are currently lacking.
Our Solutions for Nucleic Acid-Based PROTAC Development
Nucleic acid-based PROTACs—whether DNA-, RNA-, or aptamer-linked—are redefining the boundaries of targeted protein degradation by offering unique selectivity, programmable designs, and access to previously unreachable targets. At BOC Sciences, we provide a full suite of end-to-end solutions designed specifically for RNA-based PROTACs, DNA-templated degraders, and hybrid oligonucleotide-PROTAC constructs. Our scientific team combines deep expertise in nucleic acid chemistry with advanced molecular degradation strategies to help you turn cutting-edge concepts into validated drug candidates.
Customized Nucleic Acid PROTAC Design Services
Our custom design services are built to support early-stage concept development through to lead optimization for nucleic acid-based PROTACs. Whether you're working with RNA aptamers, DNA-binding motifs, or CRISPR-guided elements, we offer:
- Target feasibility assessments to determine if a nucleic acid-based approach is optimal for your protein of interest.
- Design and synthesis of functionalized oligonucleotides, including linker-modified RNA, DNA, or LNA molecules with precise chemical properties.
- Integration of E3 ligase recruiters with nucleic acid scaffolds for optimal ternary complex formation.
- Structure-activity relationship (SAR) studies to refine degradation potency and selectivity.
- Custom conjugation chemistry for nucleic acid-ligand hybridization, with scalable synthesis options.
Every project is supported by experienced PhD-level scientists and guided by your unique research goals. Whether you're exploring RNA-based degrader proof-of-concept or optimizing delivery and stability, we deliver solutions tailored to your innovation pipeline.
Advanced Screening and Testing Tools for RNA and DNA-Based PROTACs
To support the validation and functional assessment of your nucleic acid-based PROTACs, we offer a suite of cutting-edge screening platforms designed for high-throughput, quantitative, and mechanistic evaluation:
- In vitro degradation assays tailored for nucleic acid chimeras to measure target protein loss across diverse cell types.
- Fluorescent and luminescent reporter assays to monitor degradation kinetics in real time.
- RNA-target and DNA-templated target engagement studies using biophysical tools such as SPR, MST, and ITC.
- CRISPR-compatible cellular models to assess degradation in gene-edited or knock-in systems.
- Stability and biodistribution profiling for RNA- or DNA-conjugated PROTACs in biological matrices.
By leveraging our validated workflows and custom assay development capabilities, you can accelerate lead selection, optimize delivery, and de-risk your nucleic acid-based degrader programs early in the development process.
Developing nucleic acid-based PROTACs? We specialize in RNA/DNA conjugation chemistry, linker optimization, and delivery systems tailored for nucleic acid degraders. Let us help you bring your next-generation PROTACs from concept to reality.
References
- Image retrieved from Figure 1 " Nucleic acids as POI ligands in PROTACs," Luo H.; et al., used under [CC BY 4.0](https://creativecommons.org/licenses/by/4.0/). The original image was not modified.
- Luo H, Tian Y, Abdullah R, et al. Advancing Design Strategy of PROTACs for Cancer Therapy[J]. MedComm, 2025, 6(7): e70258. https://doi.org/10.1002/mco2.70258.
- Cecchini C, Pannilunghi S, Tardy S, et al. From conception to development: investigating PROTACs features for improved cell permeability and successful protein degradation[J]. Frontiers in Chemistry, 2021, 9: 672267. https://doi.org/10.3389/fchem.2021.672267.
- He Y, Khan S, Huo Z, et al. Proteolysis targeting chimeras (PROTACs) are emerging therapeutics for hematologic malignancies[J]. Journal of Hematology & Oncology, 2020, 13(1): 103. https://doi.org/10.1186/s13045-020-00924-z.
- Sincere N I, Anand K, Ashique S, et al. PROTACs: emerging targeted protein degradation approaches for advanced druggable strategies[J]. Molecules, 2023, 28(10): 4014. https://doi.org/10.3390/molecules28104014.

 Fig. 1 Nucleic acids as POI ligands in PROTACs.1,2
Fig. 1 Nucleic acids as POI ligands in PROTACs.1,2