Chemically induced target protein degradation is a new strategy to regulate the level of intracellular protein. Bifunctional molecule that binds to the target protein and E3 ubiquitin ligase is designed to induce protein degradation, and the Proteolysis-targeting chimeras (Protac®) is a molecule designed to achieve this goal. By connecting the target molecule with E3 ubiquitin ligase, the target protein is degraded after ubiquitination.
Because the molecular weight of Protac® molecules is significantly higher than that of traditional drugs, the permeability of Protacs molecules plays an important role in the activity of these molecules. An article published in ACS Medicinal Chemistry Letters in June 2020 systematically discussed the relationship among cellular permeability of Protacs molecule, ligand, and linker structure.
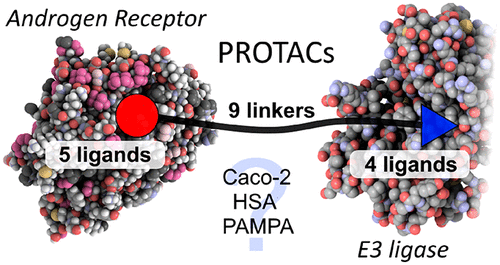 Fig.1. Androgen receptor Protacs molecules [1]
Fig.1. Androgen receptor Protacs molecules [1]
Design of Protacs
Androgen receptor (AR) is a nuclear hormone receptor for DNA transcription factor, which mediates the expression of growth factors and cell signal-dependent genes by binding to the endogenous ligand testosterone. The ligand-binding domain (LBD) on AR provides a clear binding pocket for endogenous steroid ligands. At present, many selective androgen receptor modulators (SARMs) and Protacs targeting androgen receptor (AR) have been reported. On the basis of these studies, , researchers chose androgen receptor (AR) as the model system. When designing Protacs, three components of such molecules need to be considered: AR ligand, E3 ubiquitin ligase ligand, and linker. In order to select a suitable ligand for AR, researchers first used the multi-parameter optimization (MPO) algorithm to determine that SARMs have good central nervous system (CNS) permeability, docked SARMs1-5 with AR's LBD, and found that SARMs1-5 (figure 2B) has a position far from the AR binding site, which was conducive to the successful use of this site by previous AR Protacs in linker.
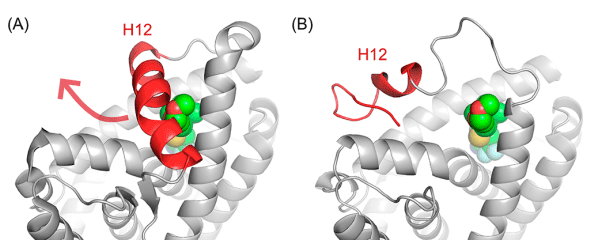 Fig. 2. Schematic diagram of molecular docking between (A) AR and 2; (B) AR ligand SARMs1-5 [1]
Fig. 2. Schematic diagram of molecular docking between (A) AR and 2; (B) AR ligand SARMs1-5 [1]
Analysis on the eutectic structure of SARMs and AR shows that the ligand is completely wrapped in the protein helix 12 (H12) (Fig. 3A). In order for the AR ligand of Protacs to successfully connect with the linker, the protein must undergo conformational change. Previous studies have shown that H12 forms a “lid” on SARMs, and this structure can be moved to accommodate extended AR ligands. When estrogen receptor (ER) binds to antagonists, the conformation of H12 changes dramatically, so the researchers built an AR homologous model with open conformation of H12 based on the open ER protein complex of H12 (Fig. 3B). The open conformation of H12 in the model allows the linker portion of Protacs to be accommodated, giving it the opportunity to extend from the ligand binding domain to the solvent region.
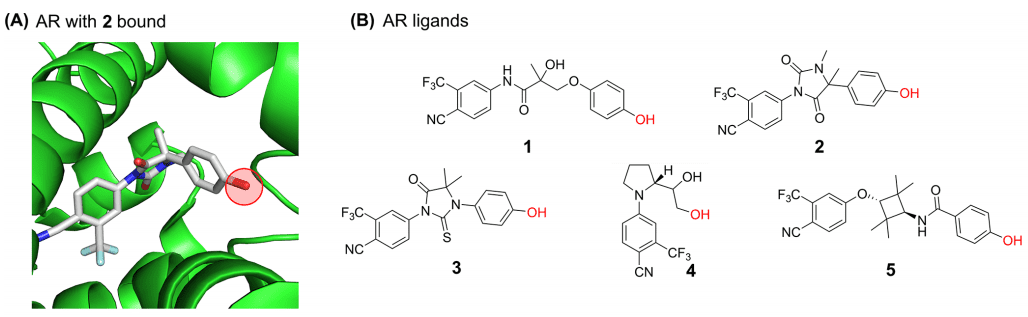 Fig. 3. Eutectic structure of (A) AR protein (AR homologous model of PDB:2PNU); (B) with open conformation H12 [1]
Fig. 3. Eutectic structure of (A) AR protein (AR homologous model of PDB:2PNU); (B) with open conformation H12 [1]
At present, the types of E3 ligase ligands used in Protacs molecules are limited, and researchers have selected four protein hydrolysis targeting structures with different physical and chemical properties (see structure 6-9 in Fig. 4C). Among them, 6 and 7 are analogues of the immunomodulatory drug thalidomide, and can recruit E3 ligase Cereblon (Fig. 4D), 9 is a small ligand of VHL E3 ligase, hydrophobic amantadine 8 can recognized by the Hsp70/CHIP system when it is present on the surface of AR, resulting in AR clearance.
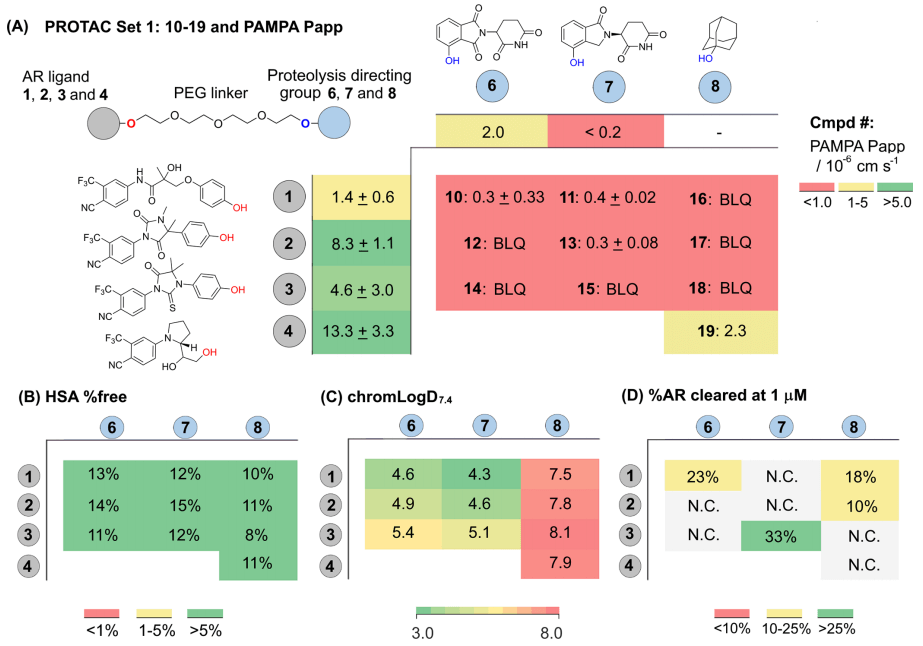 Fig. 4. (C) The targeted structure of protein hydrolysis. The linker junction is marked with blue to show the binding diagram of; (D) Schematic diagram of the combination of lenalidomide and Cereblon; (E) Schematic diagram of the combination of lenalidomide and Cereblon [1]
Fig. 4. (C) The targeted structure of protein hydrolysis. The linker junction is marked with blue to show the binding diagram of; (D) Schematic diagram of the combination of lenalidomide and Cereblon; (E) Schematic diagram of the combination of lenalidomide and Cereblon [1]
The researchers first linked AR ligands and smaller E3 ligands through PEG to synthesize a group of Protacs (Fig. 4E Protac® Set 1). They calculated logD prediction permeability and evaluated the effect of changes in the two types of ligands on Protacs permeability when the linker is constant, and then synthesized another group of Protacs using VHL ligand 9 (Fig. 4 Protac® Set 2) to evaluate the effect of changing linker on permeability and functional activity. It is worth mentioning that they introduced fluorine atoms into linker, which is the first reported fluorine-containing linker in Protacs research.
Evaluation of permeability and bioactivity of Protacs
Next, one group of Protacs were selected to evaluate their cell activity, passive permeability (PAMPA), human serum albumin (HSA) binding and chromLogD7.4. Protac® 10-19 (Fig.5) has minimized molecular weight and topological polar surface area (TPSA). Such physical and chemical properties are not common in other Protacs because their AR ligands are small and effective, the E3 ligase ligands have low molecular weight, and the PEG linkers have typical reasonable length and polar atom distribution. As shown in Fig. 5A, the PAMPA Papp detection of the AR ligand shows a wide range of permeability: ligand 1 has low permeability and ligand 4 has high permeability. According to the Protac® activity reported in the literature, the permeability of this group of Protacs may be reasonable, especially when binding the most permeable AR ligands and E3 ligase ligands. However, the analysis of PAMPA shows that the permeability of Protac® molecules is very low, even lower than the quantitative limit (Fig. 5A and Table 2). Obviously, the inherent high molecular weight and high TPSA of Protac® molecules are the main reasons for their poor permeability. However, Protac® 19, which combines 4 with adamantyl deoxynucleotides 8, has the highest PAMPA permeability in bifunctional molecular tests. LogD with higher lipophilic adamantane analogues may be beneficial to permeability (Fig. 5C).
In view of the low passive permeability measured in the PAMPA test, the bidirectional permeability of some Protac® molecules in Caco-2 cells was also tested (tables 1 and 2). The biological correlation of polarized Caco-2 monolayer cells is stronger, and the permeability in two opposite directions ("A2B" and "B2A") can be evaluated. The active transport can also be evaluated from the ratio of directional permeability.
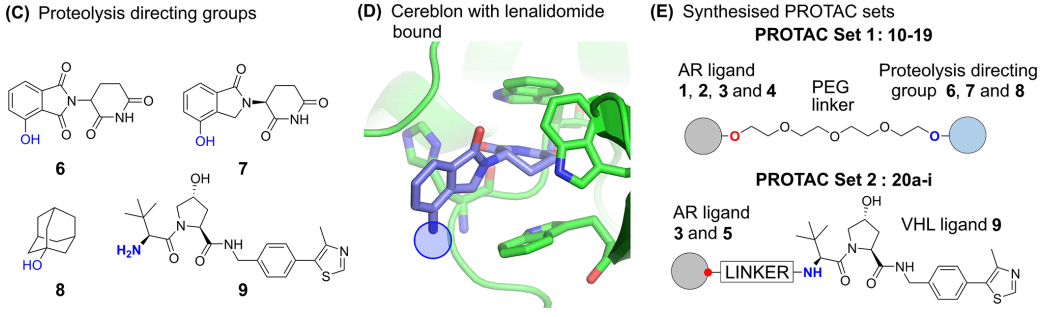 Fig.5. Structure, physiological characteristics and biological activity of Protac®
Fig.5. Structure, physiological characteristics and biological activity of Protac®
Table 1. Summary of PAMPA and Caco-2 permeability of Protacs with different E3 ligase ligands [1]
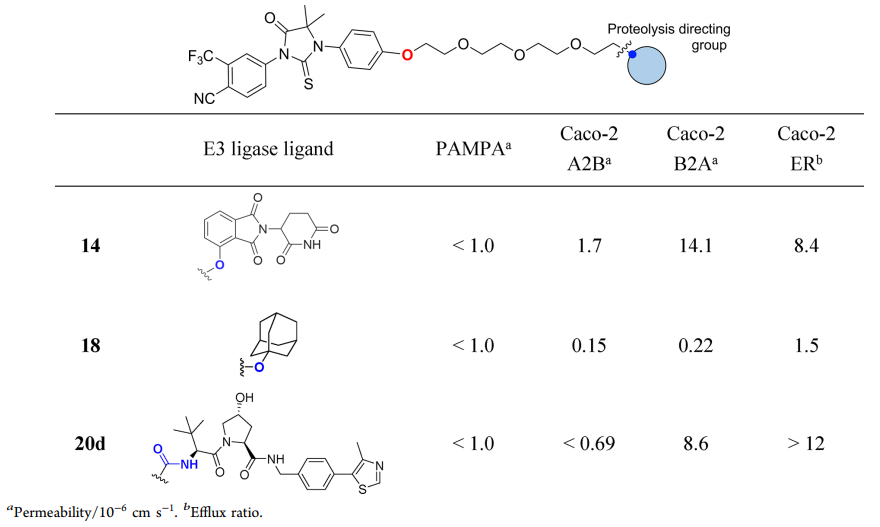
The effects of Protac® molecules with AR ligand 3 and PEG linker can be compared and identified by Caco-2 data. Among them, Protac® 14 containing Cereblon ligand has the best A2B permeability and a high B2A ratio, indicating the existence of transporter outflow (outflow ratio ER=8.4). Replacing the Cereblon ligands of Protac® with small, hydrophobic adamantane degradation products resulted in a 10-fold reduction in A2B and a 64-fold reduction in B2A, without the possibility of transporter outflow (ER=1.5). The use of larger and more polar VHL ligands in Protac® 20d directly caused the lower permeability of A2B compared to its quantitative limit. But when the ratio of B2A is 9.6×10-6cm s-1, there was an obvious outflow (ER > 12).
Table 2. Structure, physiological characteristics and permeability of targeted VHL Protac® [1]
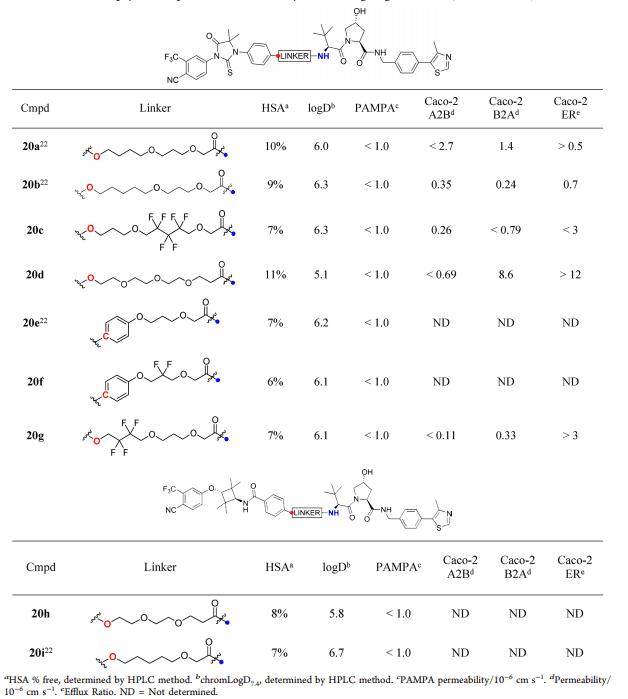
Next, the researchers used the second group of Protacs (containing VHL ligands) to explore the effect of linker on Caco-2 permeability (Table 2). It was found that the A2B of all Protacs is very low (< 1.0×10-6 cm s-1) or could not be measured at all. In addition, the linkers of Protac® 20b, 20c and 20g have different numbers of fluorine atoms, which leads to the same permeability of Protacs and low ratio of A2B to B2A. In contrast, Protac® 20d is very sensitive to the change of PEG linker, and B2A increased significantly (8.6×10-6 cm s-1), and the outflow ratio also increased. The above results indicated that the passive permeability of Protacs molecules is usually low, but the structure of linker and E3 ligase ligands has a significant effect on the efflux rate of A2B and B2A in Caco-2 cells. In addition, the binding rate of all Protacs to HSA protein is low, only 6%~15% (Fig. 4B and Table 2). In order to explain this phenomenon, more studies are needed.
Androgen-dependent human prostate cancer (LNCaP) cell line is a typical model of AR expression, which is often used in AR Protacs study. Western blotting can be used to evaluate the down-regulation of endogenous AR expression by Protacs in LNCaP cells. In order to achieve effective clearance, a stable ternary complex must be formed, and the lysine on the surface of the target protein must be presented in the correct way to achieve effective ubiquitin transfer. In general, Protac® activity can be described by two data: DCMAX and DC50. The former is the maximum percentage of protein clearance, and the latter is the concentration required to reach half of DCMAX. As shown in figure 5A, Protac® 20a was selected as the positive control, and it was found that most of the Protacs showed weak scavenging effect (DCMAX<30%) or no scavenging effect in cell experiments. In order to rule out that the linker junction of Protacs may block the binding of ligands to AR and thus lead to poor activity, the researchers tested the binding of representative Protac® 20a and 20i to AR using radioligand replacement analysis. It was found that the affinity of the two kinds of Protac® to AR is very low, although their AR ligands and linker are different, and VHL also bounds to the ligands. This is consistent with the docking results (Fig. 3B), and also shows that the binding of Protacs linker to AR ligands has no effect on its binding to the target protein in this study. However, the binding of linker to VHL ligands has been proved to be unaffected in other studies.
As shown in Fig. 5B, the analysis of the activity data of Protac® 14-15 shows that small changes in Protacs can lead to great changes in cell activity. Protac® 15 is active (DCMAX=33%, DC50=10 nm), but Protac® 14 is not active, although 14 has the highest A2B permeability and high efflux. The only structural difference between them lies in whether the ligand and protein binding part of E3 ligase contains carbonyl group. Interestingly, other research teams also observed huge differences in activity caused by carbonyl groups when studying BET Protacs[2].
Conclusion
In short, using PAMPA and Caco-2 to measure the permeability of Protac® is not an ideal method, even for the simplest Protac® that consists of medium-sized linker and relatively small AR ligands and E3 ligase ligands, although Caco-2 permeability data can relatively better reveal the permeability of Protacs. The permeability data in this paper support a catalytic model, that is, the sub-stoichiometric level of Protacs can catalyze the clearance of the target protein population. For molecules such as Protacs, there should be a minimum permeability threshold, which should be much lower than other small molecules that conform to the five rules of drugs. In addition, the chemical stability of Protacs in cells is also important for the continuous degradation of proteins.
To design Protacs, it is important to understand the cellular uptake, time stability, and kinetic mechanism of protein clearance of Protacs. A better understanding of the relationship between the Protacs structure and cell efficacy is conducive to the rational design of better molecules and promotes the clinical application of Protacs molecules.
References
- Scott, D. E., Rooney, T. P., Bayle, E. D., Mirza, T., Willems, H. M., Clarke, J. H., ... & Skidmore, J. (2020). Systematic Investigation of the Permeability of Androgen Receptor Protacs. ACS Medicinal Chemistry Letters.
- Qin, C., Hu, Y., Zhou, B., Fernandez-Salas, E., Yang, C. Y., Liu, L., ... & Meagher, J. (2018). Discovery of QCA570 as an exceptionally potent and efficacious proteolysis targeting chimera (Protac®) degrader of the bromodomain and extra-terminal (BET) proteins capable of inducing complete and durable tumor regression. Journal of medicinal chemistry, 61(15), 6685-6704.

 Fig.1. Androgen receptor Protacs molecules [1]
Fig.1. Androgen receptor Protacs molecules [1] Fig. 2. Schematic diagram of molecular docking between (A) AR and 2; (B) AR ligand SARMs1-5 [1]
Fig. 2. Schematic diagram of molecular docking between (A) AR and 2; (B) AR ligand SARMs1-5 [1] Fig. 3. Eutectic structure of (A) AR protein (AR homologous model of PDB:2PNU); (B) with open conformation H12 [1]
Fig. 3. Eutectic structure of (A) AR protein (AR homologous model of PDB:2PNU); (B) with open conformation H12 [1] Fig. 4. (C) The targeted structure of protein hydrolysis. The linker junction is marked with blue to show the binding diagram of; (D) Schematic diagram of the combination of lenalidomide and Cereblon; (E) Schematic diagram of the combination of lenalidomide and Cereblon [1]
Fig. 4. (C) The targeted structure of protein hydrolysis. The linker junction is marked with blue to show the binding diagram of; (D) Schematic diagram of the combination of lenalidomide and Cereblon; (E) Schematic diagram of the combination of lenalidomide and Cereblon [1] Fig.5. Structure, physiological characteristics and biological activity of Protac®
Fig.5. Structure, physiological characteristics and biological activity of Protac®

