While In recent years, protein degraders such as PROTAC have achieved a number of milestones and attracted much attention from both scientific and industrial communities in recent years, DUBTAC, a class of protein stabilization technology opposite to PROTAC, has also seen a breakthrough.
OTUB1 is a member of the deubiquitylating enzymes (DUBS) OUT family. EN523 is a covalent ligand of OTUB1. Since EN523 covalently interacts with the allosteric cysteine residue Cys23 of OTUB1 and does not interfere with catalytic residue Cys91 function, the enzymatic activity function of OTUB1 is not affected after binding. This discovery leads to the development of a deubiquitinase-targeting chimera (DUBTAC) that stabilizes CTFR and holds potential for the treatment of cystic fibrosis.
Building on previous work, Wenyi Wei's group at Harvard Medical School has developed a universal transcription factor (TF)-DUBTAC platform that selectively stabilizes the tumor suppressor TF. The study was published recently in the Journal of the American Chemical Society.
The structure of the TF-DUBTAC technique consists of two parts. DUBL-X-BCN combines OTUB1 ligand EN523 with bicyclo[6.1.0]non-4-yne (BCN) by linker groups of different chain lengths, and can be coupled to a DNA oligomer modified with azide group at the 5’ end (N3-ODN, bound to TF) after Strain-promoted alkyne-azide cycloadditions (SPAAC), forming TF-DUBTAC that functions as a stable tumor suppressor.
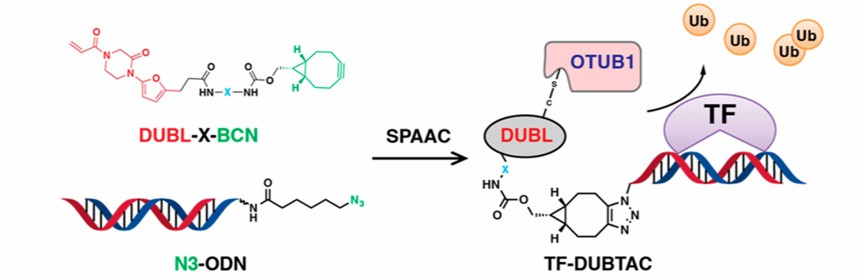 Fig 1. The mechanism of TF-DUBTAC (Liu, 2022)
Fig 1. The mechanism of TF-DUBTAC (Liu, 2022)
FOXO3A can exert a significant tumor suppressive effect on Akt, IκB and other signaling pathways, so researchers first select FOXO3A as the first TF of TF-DUBTAC platform. FOXO-DUBTAC with a chain length of 6-8 methylene groups is found to produce the best with FOXO3A through SPAAC, and the researchers chose FOXO-DUBTAC #6 with a methylene group of 7 for further cell experiments. FOXO-DUBTACs #6 stabilized FOXO3A levels in HeLa cells in a concentration-dependent manner, whereas FOXO1, FOXO3A-like TF, levels were not affected. According to the researchers, this specificity may arise from the following aspects:
- Structures other than the dna-binding pocket may affect the formation of the otub1-dubtac-foxo1 ternary complex;
- The distance between OTUB1 and the polyubiquitin chain on FOXO1 is not conducive to ubiquitin removal.
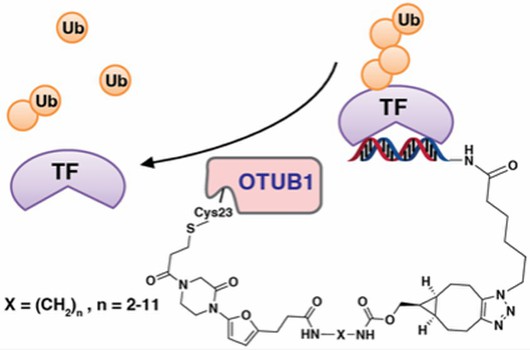 Fig 2. Removal of TF polyubiquitin by TF-DubTAC (Liu, 2022)
Fig 2. Removal of TF polyubiquitin by TF-DubTAC (Liu, 2022)
The effect of FOXO-DUbTA #6 on FOXO3A downstream signaling targets was further determined. The results showed that FOXO3A promoted p27 (inhibition of cell division) and BIM (proapoptosis) levels increased in a dose-dependent manner and decreased MYC (promotion of tumor immune escape) levels.
Mechanistic study showed that FOXO-DUBTAC #6 increased the levels of FOXO3A, p27 and BIM in OTUB1+/+ cells, but not in OTUB1-/- cells. FOXO-DUBTAC #6 inhibited tumorigenesis in wild-type HeLa cells but was unable to prevent this process in OTUB1-KO cells. Thus, these results suggest that the stabilizing effect of FOXO-DUBTAC #6 on FOXO3A is OTUB1-dependent.
To evaluate the generality of the TF-DUBTAC platform, TF-DUBTAC activity was evaluated in two other types of p53 and IRF3 with well-defined DNA-binding domains. The results showed that the corresponding TF-DUBTAC stabilized intracellular p53 and IRF3, respectively, in an OTUB1-dependent manner.
In summary, the investigators have developed a TF-targeting stabilization technique that can be used for tumor suppression. Currently, small molecule drugs targeting TF are lacking due to their "non-druggability". Since the DNA-binding domains of the vast majority of TFs are known (90% of the more than 1500 TFs have been identified so far), the TF-DUBTAC technology is expected to serve as a general platform for activation and stabilization of TFS.
It should be noted that the lower specificity of TF-DUBTAC relative to TF-PROTAC previously proposed by this experimental group may lead to non-specific stabilization, and the reasons may include:
- Many TFs have similar dna-binding domains, and high doses of TF-DUBTAC used may produce non-specific binding;
- After the target TF forms complexes with other proteins, TF-DUbTAC may remove polyubiquitin chains from the complex proteins, resulting in off-target stabilization. Therefore, it is necessary to further improve the affinity and specificity of TF-DUBTAC.
Reference
- Jing Liu et al. TF-DUBTACs Stabilize Tumor Suppressor Transcription Factors. Journal of the American Chemical Society. 2022.

 Fig 1. The mechanism of TF-DUBTAC (Liu, 2022)
Fig 1. The mechanism of TF-DUBTAC (Liu, 2022) Fig 2. Removal of TF polyubiquitin by TF-DubTAC (Liu, 2022)
Fig 2. Removal of TF polyubiquitin by TF-DubTAC (Liu, 2022)